Bayesian estimation of the prevalence of antimicrobial resistance: a mathematical modelling study
- PMID: 39051678
- PMCID: PMC11368424
- DOI: 10.1093/jac/dkae230
Bayesian estimation of the prevalence of antimicrobial resistance: a mathematical modelling study
Abstract
Background: Estimates of the prevalence of antimicrobial resistance (AMR) underpin effective antimicrobial stewardship, infection prevention and control, and optimal deployment of antimicrobial agents. Typically, the prevalence of AMR is determined from real-world antimicrobial susceptibility data that are time delimited, sparse, and often biased, potentially resulting in harmful and wasteful decision-making. Frequentist methods are resource intensive because they rely on large datasets.
Objectives: To determine whether a Bayesian approach could present a more reliable and more resource-efficient way to estimate population prevalence of AMR than traditional frequentist methods.
Methods: Retrospectively collected, open-source, real-world pseudonymized healthcare data were used to develop a Bayesian approach for estimating the prevalence of AMR by combination with prior AMR information from a contextualized review of literature. Iterative random sampling and cross-validation were used to assess the predictive accuracy and potential resource efficiency of the Bayesian approach compared with a standard frequentist approach.
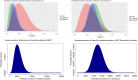
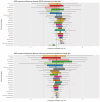
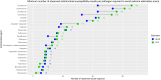
Results: Bayesian estimation of AMR prevalence made fewer extreme estimation errors than a frequentist estimation approach [n = 74 (6.4%) versus n = 136 (11.8%)] and required fewer observed antimicrobial susceptibility results per pathogen on average [mean = 28.8 (SD = 22.1) versus mean = 34.4 (SD = 30.1)] to avoid any extreme estimation errors in 50 iterations of the cross-validation. The Bayesian approach was maximally effective and efficient for drug-pathogen combinations where the actual prevalence of resistance was not close to 0% or 100%.
Conclusions: Bayesian estimation of the prevalence of AMR could provide a simple, resource-efficient approach to better inform population infection management where uncertainty about AMR prevalence is high.
© The Author(s) 2024. Published by Oxford University Press on behalf of British Society for Antimicrobial Chemotherapy.
Figures


References
-
- Tipping ME. Bayesian Inference: an introduction to principles and practice in machine learning. In: Bousquet O, von Luxburg U, Rätsch G, eds. Advanced Lectures on Machine Learning: ML 2003. Lecture Notes in Computer Science (vol 3176). Springer, 2004; 41–62.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

