CDK9 Inhibition by Dinaciclib Is a Therapeutic Vulnerability in Epithelioid Hemangioendothelioma
- PMID: 39052240
- PMCID: PMC11398983
- DOI: 10.1158/1078-0432.CCR-24-1097
CDK9 Inhibition by Dinaciclib Is a Therapeutic Vulnerability in Epithelioid Hemangioendothelioma
Abstract
Purpose: There are no effective treatment options for patients with aggressive epithelioid hemangioendothelioma (EHE) driven by the TAZ-CAMTA1 (TC) fusion gene. Here, we aimed to understand the regulation of TC using pharmacologic tools and identify vulnerabilities that can potentially be exploited for the treatment of EHE.
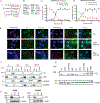
Experimental design: TC is a transcriptional coregulator; we hypothesized that compounds that reduce TC nuclear levels, either through translocation of TC to the cytoplasm, or through degradation, would render TC less oncogenic. TC localization was monitored using immunofluorescence in an EHE tumor cell line. Two target-selective libraries were used to identify small molecules that reduce TC localization in the nucleus. The ability of the shortlisted hits to affect cell viability, apoptosis, and tumorigenesis was also evaluated.
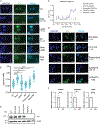
Results: Basal TC remained "immobile" in the nucleus; administration of cyclin-dependent kinase (CDK) inhibitors such as CGP60474 and dinaciclib (Dina) mobilized TC. "Mobile" TC shuttled between the nucleus and cytoplasm; however, it was eventually degraded through proteasomes. This dramatically suppressed the levels of TC-regulated transcripts and cell viability, promoted apoptosis, and reduced the area of metastatic lesions in the allograft model of EHE. We specifically identified that the inhibition of CDK9, a transcriptional CDK, destabilizes TC.
Conclusions: The CDK inhibitor Dina exhibited antitumorigenic properties both in vitro and in vivo in EHE models. Dina has been rigorously tested in clinical trials and displayed an acceptable toxicity profile. Therefore, there is a potential therapeutic window for repurposing Dina for the treatment of EHE.
©2024 American Association for Cancer Research.
Conflict of interest statement
Figures



References
-
- Tanas MR, Sboner A, Oliveira AM, Erickson-Johnson MR, Hespelt J, Hanwright PJ, et al. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med. 2011;3(98):98ra82. - PubMed
-
- Dermawan JK, Azzato EM, Billings SD, Fritchie KJ, Aubert S, Bahrami A, et al. YAP1-TFE3-fused hemangioendothelioma: a multi-institutional clinicopathologic study of 24 genetically-confirmed cases. Mod Pathol. 2021;34(12):2211–21. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

