The genomic landscape of lung cancer in never-smokers from the Women's Health Initiative
- PMID: 39052387
- PMCID: PMC11385083
- DOI: 10.1172/jci.insight.174643
The genomic landscape of lung cancer in never-smokers from the Women's Health Initiative
Abstract
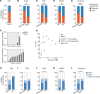
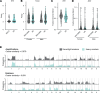
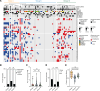
Over 200,000 individuals are diagnosed with lung cancer in the United States every year, with a growing proportion of cases, especially lung adenocarcinoma, occurring in individuals who have never smoked. Women over the age of 50 comprise the largest affected demographic. To understand the genomic drivers of lung adenocarcinoma and therapeutic response in this population, we performed whole genome and/or whole exome sequencing on 73 matched lung tumor/normal pairs from postmenopausal women who participated in the Women's Health Initiative. Somatic copy number alterations showed little variation by smoking status, suggesting that aneuploidy may be a general characteristic of lung cancer regardless of smoke exposure. Similarly, clock-like and APOBEC mutation signatures were prevalent but did not differ in tumors from smokers and never-smokers. However, mutations in both EGFR and KRAS showed unique allelic differences determined by smoking status that are known to alter tumor response to targeted therapy. Mutations in the MYC-network member MGA were more prevalent in tumors from smokers. Fusion events in ALK, RET, and ROS1 were absent, likely due to age-related differences in fusion prevalence. Our work underscores the profound effect of smoking status, age, and sex on the tumor mutational landscape and identifies areas of unmet medical need.
Keywords: Genetic variation; Genetics; Lung cancer; Molecular diagnosis; Therapeutics.
Conflict of interest statement
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

