Spillover of highly pathogenic avian influenza H5N1 virus to dairy cattle
- PMID: 39053575
- PMCID: PMC11485258
- DOI: 10.1038/s41586-024-07849-4
Spillover of highly pathogenic avian influenza H5N1 virus to dairy cattle
Abstract
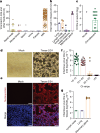
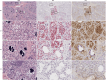
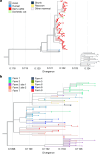
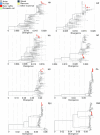
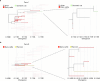
The highly pathogenic avian influenza (HPAI) H5N1 virus clade 2.3.4.4b has caused the death of millions of domestic birds and thousands of wild birds in the USA since January 2022 (refs. 1-4). Throughout this outbreak, spillovers to mammals have been frequently documented5-12. Here we report spillover of the HPAI H5N1 virus to dairy cattle across several states in the USA. The affected cows displayed clinical signs encompassing decreased feed intake, altered faecal consistency, respiratory distress and decreased milk production with abnormal milk. Infectious virus and viral RNA were consistently detected in milk from affected cows. Viral distribution in tissues via immunohistochemistry and in situ hybridization revealed a distinct tropism of the virus for the epithelial cells lining the alveoli of the mammary gland in cows. Whole viral genome sequences recovered from dairy cows, birds, domestic cats and a raccoon from affected farms indicated multidirectional interspecies transmissions. Epidemiological and genomic data revealed efficient cow-to-cow transmission after apparently healthy cows from an affected farm were transported to a premise in a different state. These results demonstrate the transmission of the HPAI H5N1 clade 2.3.4.4b virus at a non-traditional interface, underscoring the ability of the virus to cross species barriers.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
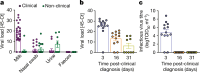








References
-
- Youk, S. et al. H5N1 highly pathogenic avian influenza clade 2.3.4.4b in wild and domestic birds: introductions into the United States and reassortments, December 2021–April 2022. Virology587, 109860 (2023). - PubMed
-
- USDA–APHIS. Detections of highly pathogenic avian influenza in wild birds. USDA–APHIShttps://www.aphis.usda.gov/livestock-poultry-disease/avian/avian-influen... (2024).
-
- USDA–APHIS. Confirmations of highly pathogenic avian influenza in commercial and backyard flocks. USDA–APHIShttps://www.aphis.usda.gov/livestock-poultry-disease/avian/avian-influen... (2024).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

