Immune cell topography of head and neck cancer
- PMID: 39053947
- PMCID: PMC11284952
- DOI: 10.1136/jitc-2024-009550
Immune cell topography of head and neck cancer
Abstract
Background: Approximately 50% of head and neck squamous cell carcinomas (HNSCC) recur after treatment with curative intent. Immune checkpoint inhibitors are treatment options for recurrent/metastatic HNSCC; however, less than 20% of patients respond. To increase this response rate, it is fundamental to increase our understanding of the spatial tumor immune microenvironment (TIME).
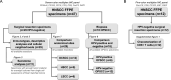
Methods: In total, 53 HNSCC specimens were included. Using a seven-color multiplex immunohistochemistry panel we identified tumor cells, CD163+macrophages, B cells, CD8+T cells, CD4+T helper cells and regulatory T cells (Tregs) in treatment-naive surgical resection specimens (n=29) and biopsies (n=18). To further characterize tumor-infiltrating CD8+T cells, we stained surgical resection specimens (n=12) with a five-color tumor-resident panel including CD103, Ki67, CD8 and pan-cytokeratin. Secretome analysis was performed on matched tumor suspensions (n=11) to measure protein levels.
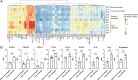
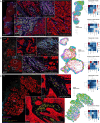
Results: Based on CD8+T cell infiltrates, we identified four different immunotypes: fully infiltrated, stroma-restricted, immune-excluded, and immune-desert. We found higher cytokine levels in fully infiltrated tumors compared with other immunotypes. While the highest immune infiltrates were observed in the invasive margin for all immune cells, CD163+macrophages and Tregs had the highest tendency to infiltrate the tumor center. Within the tumor center, especially B cells stayed at the tumor stroma, whereas CD163+macrophages, followed by T cells, were more often localized within tumor fields. Also, B cells were found further away from other cells and often formed aggregates while T cells and CD163+macrophages tended to be more closely located to each other. Across resection specimens from various anatomical sites within the head and neck, oral cavity tumors exhibited the highest densities of Tregs. Moreover, the distance from B cells and T cells to tumor cells was shortest in oral cavity squamous cell carcinoma (OCSCC), suggesting more interaction between lymphocytes and tumor cells. Also, the fraction of T cells within 10 µm of CD163+macrophages was lowest in OCSCC, indicating fewer myeloid/T-cell suppressive interactions in OCSCC.
Conclusions: We comprehensively described the TIME of HNSCC using a unique data set of resection specimens. We discovered that the composition, as well as the relative localization of immune cells in the TIME, differed in distinct anatomical sites of the head and neck.
Keywords: B cell; Head and Neck Cancer; Macrophage; T cell; Tumor microenvironment - TME.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures




References
-
- Ferris RL, Blumenschein G, Fayette J, et al. Nivolumab vs investigator’s choice in recurrent or metastatic squamous cell carcinoma of the head and neck: 2-year long-term survival update of CheckMate 141 with analyses by tumor PD-L1 expression. Oral Oncol. 2018;81:45–51. doi: 10.1016/j.oraloncology.2018.04.008. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
