TFEB/3 Govern Repair Schwann Cell Generation and Function Following Peripheral Nerve Injury
- PMID: 39054068
- PMCID: PMC11358533
- DOI: 10.1523/JNEUROSCI.0198-24.2024
TFEB/3 Govern Repair Schwann Cell Generation and Function Following Peripheral Nerve Injury
Abstract
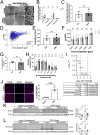
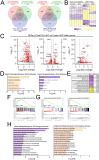
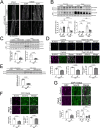
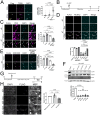
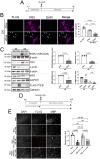
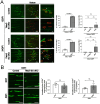
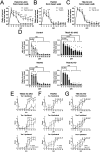
TFEB and TFE3 (TFEB/3), key regulators of lysosomal biogenesis and autophagy, play diverse roles depending on cell type. This study highlights a hitherto unrecognized role of TFEB/3 crucial for peripheral nerve repair. Specifically, they promote the generation of progenitor-like repair Schwann cells after axonal injury. In Schwann cell-specific TFEB/3 double knock-out mice of either sex, the TFEB/3 loss disrupts the transcriptomic reprogramming that is essential for the formation of repair Schwann cells. Consequently, mutant mice fail to populate the injured nerve with repair Schwann cells and exhibit defects in axon regrowth, target reinnervation, and functional recovery. TFEB/3 deficiency inhibits the expression of injury-responsive repair Schwann cell genes, despite the continued expression of c-jun, a previously identified regulator of repair Schwann cell function. TFEB/3 binding motifs are enriched in the enhancer regions of injury-responsive genes, suggesting their role in repair gene activation. Autophagy-dependent myelin breakdown is not impaired despite TFEB/3 deficiency. These findings underscore a unique role of TFEB/3 in adult Schwann cells that is required for proper peripheral nerve regeneration.
Keywords: Runx2; Schwann cell; c-Jun; myelin; regeneration; sciatic nerve injury.
Copyright © 2024 the authors.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Abercrombie M, Johnson ML (1946) Quantitative histology of Wallerian degeneration; nuclear population in rabbit sciatic nerve. J Anat 80:37–50. - PubMed
-
- Arthur-Farraj PJ, Morgan CC, Adamowicz M, Gomez-Sanchez JA, Fazal SV, Beucher A, Razzaghi B, Mirsky R, Jessen KR, Aitman TJ (2017) Changes in the coding and non-coding transcriptome and DNA methylome that define the Schwann cell repair phenotype after nerve injury. Cell Rep 20:2719–2734. 10.1016/j.celrep.2017.08.064 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
