Coding, or non-coding, that is the question
- PMID: 39054345
- PMCID: PMC11369213
- DOI: 10.1038/s41422-024-00975-8
Coding, or non-coding, that is the question
Abstract
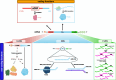
The advent of high-throughput sequencing uncovered that our genome is pervasively transcribed into RNAs that are seemingly not translated into proteins. It was also found that non-coding RNA transcripts outnumber canonical protein-coding genes. This mindboggling discovery prompted a surge in non-coding RNA research that started unraveling the functional relevance of these new genetic units, shaking the classic definition of "gene". While the non-coding RNA revolution was still taking place, polysome/ribosome profiling and mass spectrometry analyses revealed that peptides can be translated from non-canonical open reading frames. Therefore, it is becoming evident that the coding vs non-coding dichotomy is way blurrier than anticipated. In this review, we focus on several examples in which the binary classification of coding vs non-coding genes is outdated, since the same bifunctional gene expresses both coding and non-coding products. We discuss the implications of this intricate usage of transcripts in terms of molecular mechanisms of gene expression and biological outputs, which are often concordant, but can also surprisingly be discordant. Finally, we discuss the methodological caveats that are associated with the study of bifunctional genes, and we highlight the opportunities and challenges of therapeutic exploitation of this intricacy towards the development of anticancer therapies.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

