Alzheimer's disease detection using data fusion with a deep supervised encoder
- PMID: 39055313
- PMCID: PMC11271260
- DOI: 10.3389/frdem.2024.1332928
Alzheimer's disease detection using data fusion with a deep supervised encoder
Abstract
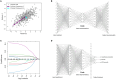
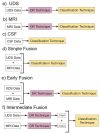
Alzheimer's disease (AD) is affecting a growing number of individuals. As a result, there is a pressing need for accurate and early diagnosis methods. This study aims to achieve this goal by developing an optimal data analysis strategy to enhance computational diagnosis. Although various modalities of AD diagnostic data are collected, past research on computational methods of AD diagnosis has mainly focused on using single-modal inputs. We hypothesize that integrating, or "fusing," various data modalities as inputs to prediction models could enhance diagnostic accuracy by offering a more comprehensive view of an individual's health profile. However, a potential challenge arises as this fusion of multiple modalities may result in significantly higher dimensional data. We hypothesize that employing suitable dimensionality reduction methods across heterogeneous modalities would not only help diagnosis models extract latent information but also enhance accuracy. Therefore, it is imperative to identify optimal strategies for both data fusion and dimensionality reduction. In this paper, we have conducted a comprehensive comparison of over 80 statistical machine learning methods, considering various classifiers, dimensionality reduction techniques, and data fusion strategies to assess our hypotheses. Specifically, we have explored three primary strategies: (1) Simple data fusion, which involves straightforward concatenation (fusion) of datasets before inputting them into a classifier; (2) Early data fusion, in which datasets are concatenated first, and then a dimensionality reduction technique is applied before feeding the resulting data into a classifier; and (3) Intermediate data fusion, in which dimensionality reduction methods are applied individually to each dataset before concatenating them to construct a classifier. For dimensionality reduction, we have explored several commonly-used techniques such as principal component analysis (PCA), autoencoder (AE), and LASSO. Additionally, we have implemented a new dimensionality-reduction method called the supervised encoder (SE), which involves slight modifications to standard deep neural networks. Our results show that SE substantially improves prediction accuracy compared to PCA, AE, and LASSO, especially in combination with intermediate fusion for multiclass diagnosis prediction.
Keywords: Alzheimer’s biomarkers; Alzheimer’s disease; data integration; diagnosis prediction; dimensionality reduction; multimodal fusion; multiview data integration.
Conflict of interest statement
Conflict of interest The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Andrews J. S., Beach T. G., Buracchio T., Carrillo M. C., Dunn B., Graf A., et al. . (2023). Revised Criteria for Diagnosis and Staging of Alzheimer's Disease: Alzheimers Association Workgroup. Available online at: https://aaic.alz.org/diagnostic-criteria.asp (accessed December 13, 2023).
-
- Beekly D. L., Ramos E. M., van Belle G., Deitrich W., Clark A. D., Jacka M. E., et al. . (2004). The national Alzheimer's coordinating center (NACC) database: an Alzheimer disease database. Alzheimer Dis. Assoc. Disor. 18, 270–277. - PubMed
Grants and funding
- P20 AG068053/AG/NIA NIH HHS/United States
- P30 AG066515/AG/NIA NIH HHS/United States
- P30 AG062421/AG/NIA NIH HHS/United States
- P30 AG066508/AG/NIA NIH HHS/United States
- P30 AG066519/AG/NIA NIH HHS/United States
- P30 AG072973/AG/NIA NIH HHS/United States
- P30 AG066462/AG/NIA NIH HHS/United States
- P30 AG066530/AG/NIA NIH HHS/United States
- P30 AG066509/AG/NIA NIH HHS/United States
- P20 AG068077/AG/NIA NIH HHS/United States
- P30 AG066546/AG/NIA NIH HHS/United States
- P30 AG072972/AG/NIA NIH HHS/United States
- P30 AG072979/AG/NIA NIH HHS/United States
- P20 AG068082/AG/NIA NIH HHS/United States
- P30 AG072975/AG/NIA NIH HHS/United States
- P30 AG066444/AG/NIA NIH HHS/United States
- P30 AG066507/AG/NIA NIH HHS/United States
- P30 AG072946/AG/NIA NIH HHS/United States
- F30 AG079610/AG/NIA NIH HHS/United States
- P30 AG066518/AG/NIA NIH HHS/United States
- P30 AG066511/AG/NIA NIH HHS/United States
- U24 AG072122/AG/NIA NIH HHS/United States
- P30 AG066512/AG/NIA NIH HHS/United States
- P30 AG072978/AG/NIA NIH HHS/United States
- P30 AG062429/AG/NIA NIH HHS/United States
- P30 AG062422/AG/NIA NIH HHS/United States
- R01 AG079280/AG/NIA NIH HHS/United States
- P30 AG072977/AG/NIA NIH HHS/United States
- P30 AG062677/AG/NIA NIH HHS/United States
- P20 AG068024/AG/NIA NIH HHS/United States
- P30 AG072958/AG/NIA NIH HHS/United States
- P30 AG062715/AG/NIA NIH HHS/United States
- P30 AG066506/AG/NIA NIH HHS/United States
- P30 AG066468/AG/NIA NIH HHS/United States
- P30 AG072976/AG/NIA NIH HHS/United States
- P30 AG072947/AG/NIA NIH HHS/United States
- P30 AG072931/AG/NIA NIH HHS/United States
- P30 AG066514/AG/NIA NIH HHS/United States
- P30 AG072959/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

