Citalopram exposure of hESCs during neuronal differentiation identifies dysregulated genes involved in neurodevelopment and depression
- PMID: 39055655
- PMCID: PMC11269147
- DOI: 10.3389/fcell.2024.1428538
Citalopram exposure of hESCs during neuronal differentiation identifies dysregulated genes involved in neurodevelopment and depression
Abstract
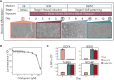
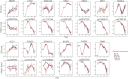
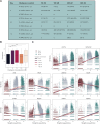
Selective serotonin reuptake inhibitors (SSRIs), including citalopram, are widely used antidepressants during pregnancy. However, the effects of prenatal exposure to citalopram on neurodevelopment remain poorly understood. We aimed to investigate the impact of citalopram exposure on early neuronal differentiation of human embryonic stem cells using a multi-omics approach. Citalopram induced time- and dose-dependent effects on gene expression and DNA methylation of genes involved in neurodevelopmental processes or linked to depression, such as BDNF, GDF11, CCL2, STC1, DDIT4 and GAD2. Single-cell RNA-sequencing analysis revealed distinct clusters of stem cells, neuronal progenitors and neuroblasts, where exposure to citalopram subtly influenced progenitor subtypes. Pseudotemporal analysis showed enhanced neuronal differentiation. Our findings suggest that citalopram exposure during early neuronal differentiation influences gene expression patterns associated with neurodevelopment and depression, providing insights into its potential neurodevelopmental impact and highlighting the importance of further research to understand the long-term consequences of prenatal SSRI exposure.
Keywords: DNA methylation; citalopram; depression; epigenetics; human embryonic stem cells; multi-omics; neurodevelopment; single-cell RNA-seq.
Copyright © 2024 Spildrejorde, Leithaug, Samara, Aass, Sharma, Acharya, Nordeng, Gervin and Lyle.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Aryee M. J., Jaffe A. E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A. P., Hansen K. D., et al. (2014). Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369. 10.1093/bioinformatics/btu049 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

