Characterizing replisome disassembly in human cells
- PMID: 39055910
- PMCID: PMC11269944
- DOI: 10.1016/j.isci.2024.110260
Characterizing replisome disassembly in human cells
Abstract
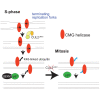
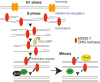
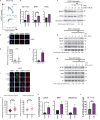
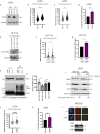
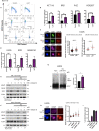
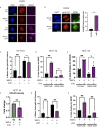
To ensure timely duplication of the entire eukaryotic genome, thousands of replication machineries (replisomes) act on genomic DNA at any time during S phase. In the final stages of this process, replisomes are unloaded from chromatin. Unloading is driven by polyubiquitylation of MCM7, a subunit of the terminated replicative helicase, and processed by p97/VCP segregase. Most of our knowledge of replication termination comes from model organisms, and little is known about how this process is executed and regulated in human somatic cells. Here we show that replisome disassembly in this system requires CUL2LRR1-driven MCM7 ubiquitylation, p97, and UBXN7 for unloading and provide evidence for "backup" mitotic replisome disassembly, demonstrating conservation of such mechanisms. Finally, we find that small-molecule inhibitors against Cullin ubiquitin ligases (CULi) and p97 (p97i) affect replisome unloading but also lead to induction of replication stress in cells, which limits their usefulness to specifically target replisome disassembly processes.
Keywords: biochemistry; cell biology; genetics.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

