SPAG7 deletion causes intrauterine growth restriction, resulting in adulthood obesity and metabolic dysfunction
- PMID: 39056292
- PMCID: PMC11281781
- DOI: 10.7554/eLife.91114
SPAG7 deletion causes intrauterine growth restriction, resulting in adulthood obesity and metabolic dysfunction
Abstract
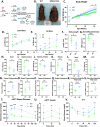
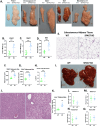
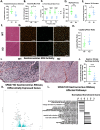
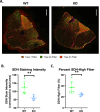
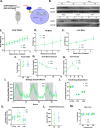
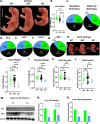
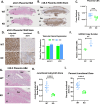
From a forward mutagenetic screen to discover mutations associated with obesity, we identified mutations in the Spag7 gene linked to metabolic dysfunction in mice. Here, we show that SPAG7 KO mice are born smaller and develop obesity and glucose intolerance in adulthood. This obesity does not stem from hyperphagia, but a decrease in energy expenditure. The KO animals also display reduced exercise tolerance and muscle function due to impaired mitochondrial function. Furthermore, SPAG7-deficiency in developing embryos leads to intrauterine growth restriction, brought on by placental insufficiency, likely due to abnormal development of the placental junctional zone. This insufficiency leads to loss of SPAG7-deficient fetuses in utero and reduced birth weights of those that survive. We hypothesize that a 'thrifty phenotype' is ingrained in SPAG7 KO animals during development that leads to adult obesity. Collectively, these results indicate that SPAG7 is essential for embryonic development and energy homeostasis later in life.
Keywords: SPAG7; developmental biology; embryonic development; insulin resistance; intrauterine growth restriction; medicine; mouse; obesity.
Plain language summary
Obesity rates are climbing worldwide, leading to an increase in associated conditions such as type 2 diabetes. While new pharmaceutical approaches are available to help individuals manage their weight, many patients do not respond to them or experience prohibitive side effects. Identifying alternative treatments will likely require pinpointing the genes and molecular actors involved in the biological processes that control weight regulation. Previous research suggests that a protein known as SPAG7 could help shape how mice use and store the energy they extract from food. Flaherty et al. therefore set out to investigate the role this protein plays in the body. To do so, they created a line of mice born without SPAG7, which they monitored closely throughout life. These animals were underweight at birth and did not eat more than other mice, yet they were obese as adults. Their ability to exercise was reduced, their muscles were weaker and contained fibers with functional defects. The mice also exhibited biological changes associated with the onset of diabetes. Yet deleting SPAG7 during adulthood led to no such changes; these mice maintained normal muscle function and body weight. Closely examining how SPAG7-deficient mice developed in the womb revealed placental defects which likely caused these animals to receive fewer nutrients from their mother. Such early-life deprivation is known to be associated with the body shifting towards maximizing its use of resources and privileging fat storage, even into and throughout adulthood. By shedding light on the biological role of SPAG7, the work by Flaherty et al. helps to better understand how developmental events can increase the likelihood of obesity later in life. Further investigations are now needed to explore whether this knowledge could help design interventions relevant to human health.
© 2023, Flaherty et al.
Conflict of interest statement
SF, LS, MP, SA, JG, AR, DS, YA, EP employee of Pfizer Inc, OB, BP, JP, YP, AO, MB, ZW was an employee of Pfizer Inc when the study was conducted, YL, RS No competing interests declared
Figures




Update of
- doi: 10.1101/2023.09.05.555087
- doi: 10.7554/eLife.91114.1
- doi: 10.7554/eLife.91114.2
References
-
- Abu-Halima M, Becker LS, Al Smadi MA, Kunz LS, Gröger L, Meese E. Expression of SPAG7 and its regulatory microRNAs in seminal plasma and seminal plasma-derived extracellular vesicles of patients with subfertility. Scientific Reports. 2023;13:3645. doi: 10.1038/s41598-023-30744-3. - DOI - PMC - PubMed
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. - DOI - PMC - PubMed
-
- Baldarelli RM, Smith CM, Finger JH, Hayamizu TF, McCright IJ, Xu J, Shaw DR, Beal JS, Blodgett O, Campbell J, Corbani LE, Frost PJ, Giannatto SC, Miers DB, Kadin JA, Richardson JE, Ringwald M. The mouse gene expression database (GXD): 2021 update. Nucleic Acids Research. 2021;49:D924–D931. doi: 10.1093/nar/gkaa914. - DOI - PMC - PubMed
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

