Single-Nucleotide Polymorphisms of TAS2R46 Affect the Receptor Downstream Calcium Regulation in Histamine-Challenged Cells
- PMID: 39056786
- PMCID: PMC11275237
- DOI: 10.3390/cells13141204
Single-Nucleotide Polymorphisms of TAS2R46 Affect the Receptor Downstream Calcium Regulation in Histamine-Challenged Cells
Abstract
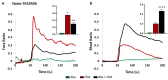
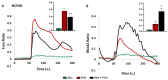
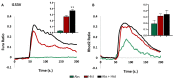
Bitter taste receptors (TAS2Rs) expressed in extraoral tissues represent a whole-body sensory system, whose role and mechanisms could be of interest for the identification of new therapeutic targets. It is known that TAS2R46s in pre-contracted airway smooth muscle cells increase mitochondrial calcium uptake, leading to bronchodilation, and that several SNPs have been identified in its gene sequence. There are very few reports on the structure-function analysis of TAS2Rs. Thus, we delved into the subject by using mutagenesis and in silico studies. We generated a cellular model that expresses native TAS2R46 to evaluate the influence of the four most common SNPs on calcium fluxes following the activation of the receptor by its specific ligand absinthin. Then, docking studies were conducted to correlate the calcium flux results to the structural mutation. The analysed SNPs differently modulate the TAS2R46 signal cascade according to the altered protein domain. In particular, the SNP in the sixth transmembrane domain of the receptors did not modulate calcium homeostasis, while the SNPs in the sequence coding for the fourth transmembrane domain completely abolished the mitochondrial calcium uptake. In conclusion, these results indicate the fourth transmembrane domain of TAS2R46 is critical for the intrinsic receptor activity.
Keywords: SNPs; TAS2R; bitter taste receptor; calcium signalling; polymorphism.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
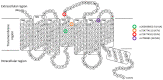




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

