KRAS Mutation Subtypes and Their Association with Other Driver Mutations in Oncogenic Pathways
- PMID: 39056802
- PMCID: PMC11274496
- DOI: 10.3390/cells13141221
KRAS Mutation Subtypes and Their Association with Other Driver Mutations in Oncogenic Pathways
Abstract
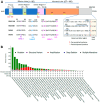
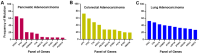
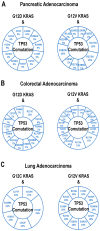
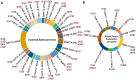
The KRAS mutation stands out as one of the most influential oncogenic mutations, which directly regulates the hallmark features of cancer and interacts with other cancer-causing driver mutations. However, there remains a lack of precise information on their cooccurrence with mutated variants of KRAS and any correlations between KRAS and other driver mutations. To enquire about this issue, we delved into cBioPortal, TCGA, UALCAN, and Uniport studies. We aimed to unravel the complexity of KRAS and its relationships with other driver mutations. We noticed that G12D and G12V are the prevalent mutated variants of KRAS and coexist with the TP53 mutation in PAAD and CRAD, while G12C and G12V coexist with LUAD. We also noticed similar observations in the case of PIK3CA and APC mutations in CRAD. At the transcript level, a positive correlation exists between KRAS and PIK3CA and between APC and KRAS in CRAD. The existence of the co-mutation of KRAS and other driver mutations could influence the signaling pathway in the neoplastic transformation. Moreover, it has immense prognostic and predictive implications, which could help in better therapeutic management to treat cancer.
Keywords: KRAS; cBioPortal; domain; mutation; predictive response; prognostic response; signaling pathway; therapeutic strategy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Tsai F.D., Lopes M.S., Zhou M., Court H., Ponce O., Fiordalisi J.J., Gierut J.J., Cox A.D., Haigis K.M., Philips M.R. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc. Natl. Acad. Sci. USA. 2015;112:779–784. doi: 10.1073/pnas.1412811112. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

