In Silico Approach: Anti-Tuberculosis Activity of Caespitate in the H37Rv Strain
- PMID: 39057029
- PMCID: PMC11275643
- DOI: 10.3390/cimb46070387
In Silico Approach: Anti-Tuberculosis Activity of Caespitate in the H37Rv Strain
Abstract
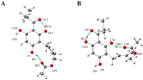
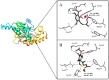
Tuberculosis is a highly lethal bacterial disease worldwide caused by Mycobacterium tuberculosis (Mtb). Caespitate is a phytochemical isolated from Helichrysum caespititium, a plant used in African traditional medicine that shows anti-tubercular activity, but its mode of action remains unknown. It is suggested that there are four potential targets in Mtb, specifically in the H37Rv strain: InhA, MabA, and UGM, enzymes involved in the formation of Mtb's cell wall, and PanK, which plays a role in cell growth. Two caespitate conformational structures from DFT conformational analysis in the gas phase (GC) and in solution with DMSO (CS) were selected. Molecular docking calculations, MM/GBSA analysis, and ADME parameter evaluations were performed. The docking results suggest that CS is the preferred caespitate conformation when interacting with PanK and UGM. In both cases, the two intramolecular hydrogen bonds characteristic of caespitate's molecular structure were maintained to achieve the most stable complexes. The MM/GBSA study confirmed that PanK/caespitate and UGM/caespitate were the most stable complexes. Caespitate showed favorable pharmacokinetic characteristics, suggesting rapid absorption, permeability, and high bioavailability. Additionally, it is proposed that caespitate may exhibit antibacterial and antimonial activity. This research lays the foundation for the design of anti-tuberculosis drugs from natural sources, especially by identifying potential drug targets in Mtb.
Keywords: H37Rv strain; MM/GBSA; antituberculosis activity; caespitate; molecular docking.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Molecular docking studies on InhA, MabA and PanK enzymes from Mycobacterium tuberculosis of ellagic acid derivatives from Ludwigia adscendens and Trewia nudiflora.In Silico Pharmacol. 2015 Dec;3(1):10. doi: 10.1186/s40203-015-0014-1. Epub 2015 Dec 8. In Silico Pharmacol. 2015. PMID: 26820895 Free PMC article.
-
In vitro and in silico evaluations of actinomycin X2and actinomycin D as potent anti-tuberculosis agents.PeerJ. 2023 Mar 8;11:e14502. doi: 10.7717/peerj.14502. eCollection 2023. PeerJ. 2023. PMID: 36935926 Free PMC article.
-
In-silico and in-vitro assessments of some fabaceae, rhamnaceae, apocynaceae, and anacardiaceae species against Mycobacterium tuberculosis H37Rv and triple-negative breast cancer cells.BMC Complement Med Ther. 2023 Jul 1;23(1):219. doi: 10.1186/s12906-023-04041-5. BMC Complement Med Ther. 2023. PMID: 37393246 Free PMC article.
-
Assessing the progress of Mycobacterium tuberculosis H37Rv structural genomics.Tuberculosis (Edinb). 2015 Mar;95(2):131-6. doi: 10.1016/j.tube.2014.12.005. Epub 2014 Dec 31. Tuberculosis (Edinb). 2015. PMID: 25578513 Review.
-
Novel targets and inhibitors of the Mycobacterium tuberculosis cytochrome bd oxidase to foster anti-tuberculosis drug discovery.Expert Opin Drug Discov. 2023 Jul-Dec;18(8):917-927. doi: 10.1080/17460441.2023.2224553. Epub 2023 Jun 18. Expert Opin Drug Discov. 2023. PMID: 37332221 Review.
Cited by
-
In Silico Screening of 1,3,4-Thiadiazole Derivatives as Inhibitors of Vascular Endothelial Growth Factor Receptor-2 (VEGFR-2).Curr Issues Mol Biol. 2024 Oct 6;46(10):11220-11235. doi: 10.3390/cimb46100666. Curr Issues Mol Biol. 2024. PMID: 39451546 Free PMC article.
References
-
- World Health Organization Global Tuberculosis Report 2020. WHO. 2020. 1, CC BY-NC-SA 3.0 IGO. [(accessed on 24 February 2024)]. Available online: https://apps.who.int/iris/handle/10665/336069.
-
- World Health Organization Global Tuberculosis Report 2019. WHO. 2019. 1, CC BY-NC-SA 3.0 IGO. [(accessed on 24 February 2024)]. Available online: https://apps.who.int/iris/handle/10665/329368.
-
- World Health Organization Global Tuberculosis Report 2018. WHO. 2018. 56, CC BY-NC-SA 3.0 IGO. [(accessed on 24 February 2024)]. Available online: https://apps.who.int/iris/handle/10665/274453.
-
- Organización Panamericana de la Salud . Tuberculosis en las Américas. Informe Regional 2021. OPS; Washington, DA, USA: 2022. CC BY-NC-SA 3.0 IGO.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

