Validation of Selected MicroRNA Transcriptome Data in the Bovine Corpus Luteum during Early Pregnancy by RT-qPCR
- PMID: 39057036
- PMCID: PMC11275921
- DOI: 10.3390/cimb46070394
Validation of Selected MicroRNA Transcriptome Data in the Bovine Corpus Luteum during Early Pregnancy by RT-qPCR
Abstract
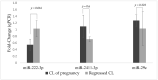
In cattle, the corpus luteum (CL) is pivotal in maintaining early pregnancy by secreting progesterone. To establish pregnancy, the conceptus produces interferon-τ, preventing luteolysis and initiating the transformation of the CL spurium into a CL verum. Although this transformation is tightly regulated, limited data are available on the expression of microRNAs (miRNAs) during and after this process. To address this gap, we re-analyzed previously published RNA-Seq data of CL from pregnant cows and regressed CL from non-pregnant cows. This analysis identified 44 differentially expressed miRNAs. From this pool, three miRNAs-bta-miR-222-3p, bta-miR-29c, and bta-miR-2411-3p-were randomly selected for relative quantification. Using bovine ovaries (n = 14) obtained from an abattoir, total RNA (including miRNAs) was extracted and converted to cDNA for RT-qPCR. The results revealed that bta-miR-222-3p was downregulated (p = 0.016) in pregnant females compared to non-pregnant cows with regressed CL. However, no differences in miRNA expression were observed between CL of pregnant and non-pregnant cows for bta-miR-29c (p > 0.32) or bta-miR-2411-3p (p > 0.60). In silico prediction approaches indicated that these miRNAs are involved in pathways regulating pregnancy maintenance, such as the VEGF- and FoxO-signaling pathways. Additionally, their biogenesis is regulated by GABPA and E2F4 transcription factors. The validation of selected miRNA expression in the CL during pregnancy by RT-qPCR provides novel insights that could potentially lead to the identification of biomarkers related to CL physiology and pregnancy outcome.
Keywords: NGS; RT-qPCR; cattle; corpus albicans; corpus luteum; microRNA; pregnancy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The Dynamics of microRNA Transcriptome in Bovine Corpus Luteum during Its Formation, Function, and Regression.Front Genet. 2017 Dec 15;8:213. doi: 10.3389/fgene.2017.00213. eCollection 2017. Front Genet. 2017. PMID: 29326752 Free PMC article.
-
Identification of microRNA transcriptome throughout the lifespan of yak (Bos grunniens) corpus luteum.Anim Biotechnol. 2023 Apr;34(2):143-155. doi: 10.1080/10495398.2021.1946552. Epub 2021 Jul 26. Anim Biotechnol. 2023. PMID: 34310260
-
Identification of stable genes in the corpus luteum of lactating Holstein cows in pregnancy and luteolysis: Implications for selection of reverse-transcription quantitative PCR reference genes.J Dairy Sci. 2020 May;103(5):4846-4857. doi: 10.3168/jds.2019-17526. Epub 2020 Mar 27. J Dairy Sci. 2020. PMID: 32229123
-
Interferon-Tau regulates a plethora of functions in the corpus luteum.Domest Anim Endocrinol. 2022 Jan;78:106671. doi: 10.1016/j.domaniend.2021.106671. Epub 2021 Aug 16. Domest Anim Endocrinol. 2022. PMID: 34509740 Review.
-
Review: Maintenance of the ruminant corpus luteum during pregnancy: interferon-tau and beyond.Animal. 2023 May;17 Suppl 1:100827. doi: 10.1016/j.animal.2023.100827. Animal. 2023. PMID: 37567676 Review.
References
-
- Tomac J., Cekinovic Đ., Aparovic J. Biology of the corpus luteum. [(accessed on 23 June 2024)];Period. Biol. 2011 113:43–49. Available online: https://hrcak.srce.hr/67235.
-
- Hansen T.R., Bott R., Romero J., Antoniazzi A., Davis J.S. Corpus Luteum and Early Pregnancy in Ruminants. In: Meidan R., editor. The Life Cycle of the Corpus Luteum. 1st ed. Springer; Cham, Switzerland: 2017. pp. 205–225.
-
- Wiltbank M.C., Meidan R., Ochoa J., Baez G.M., Giordano J.O., Ferreira J.C.P., Sartori R. Maintenance or regression of the corpus luteum during multiple decisive periods of bovine pregnancy. Anim. Reprod. 2016;13:217–233. doi: 10.21451/1984-3143-AR865. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

