Comparison of the Proteome of Huh7 Cells Transfected with Hepatitis B Virus Subgenotype A1, with or without G1862T
- PMID: 39057060
- PMCID: PMC11275860
- DOI: 10.3390/cimb46070419
Comparison of the Proteome of Huh7 Cells Transfected with Hepatitis B Virus Subgenotype A1, with or without G1862T
Abstract
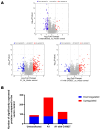
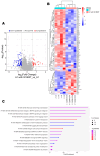
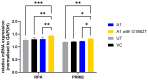
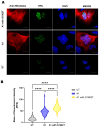
HBeAg is a non-structural, secreted protein of hepatitis B virus (HBV). Its p25 precursor is post-translationally modified in the endoplasmic reticulum. The G1862T precore mutation leads to the accumulation of P25 in the endoplasmic reticulum and activation of unfolded protein response. Using mass spectrometry, comparative proteome profiling of Huh-7 cells transfected with wildtype (WT) or G1862T revealed significantly differentially expressed proteins resulting in 12 dysregulated pathways unique to WT-transfected cells and 7 shared between cells transfected with either WT or G1862T. Except for the p38 MAPK signalling pathway, WT showed a higher number of DEPs than G1862T-transfected cells in all remaining six shared pathways. Two signalling pathways: oxidative stress and cell cycle signalling were differentially expressed only in cells transfected with G1862T. Fifteen pathways were dysregulated in G1862T-transfected cells compared to WT. The 15 dysregulated pathways were involved in the following processes: MAPK signalling, DNA synthesis and methylation, and extracellular matrix organization. Moreover, proteins involved in DNA synthesis signalling (replication protein A (RPA) and DNA primase (PRIM2)) were significantly upregulated in G1862T compared to WT. This upregulation was confirmed by mRNA quantification of both genes and immunofluorescent confocal microscopy for RPA only. The dysregulation of the pathways involved in these processes may lead to immune evasion, persistence, and uncontrolled proliferation, which are hallmarks of cancer.
Keywords: DNA primase (PRIM2); G1862T; HBV; p38 MAPK; replication protein A (RPA); subgenotype A1.
Conflict of interest statement
S.S. has a commercial interest in ReSyn Biosciences which supplied the magnetic microspheres used during mass spectrometry sample preparation. S.S. is also employed by Evosep Biosystems. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- WHO . Global Hepatitis Report 2024: Action for Access in Low- and Middle-Income Countries. WHO; Geneva, Switzerland: 2024.
-
- Chauhan R., Kazim S.N., Bhattacharjee J., Sakhuja P., Sarin S.K. Basal core promoter, precore region mutations of HBV and their association with e antigen, genotype, and severity of liver disease in patients with chronic hepatitis B in India. J. Med. Virol. 2006;78:1047–1054. doi: 10.1002/jmv.20661. - DOI - PubMed
-
- Yuen M.F., Sablon E., Yuan H.J., Hui C.K., Wong D.K.H., Doutreloigne J., Wong B.C.Y., Chan A.O.O., Lai C.L. Relationship between the development of precore and core promoter mutations and hepatitis B e antigen seroconversion in patients with chronic hepatitis B virus. J. Infect. Dis. 2002;186:1335–1338. doi: 10.1086/344327. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

