MARK2/MARK3 Kinases Are Catalytic Codependencies of YAP/TAZ in Human Cancer
- PMID: 39058094
- PMCID: PMC11609825
- DOI: 10.1158/2159-8290.CD-23-1529
MARK2/MARK3 Kinases Are Catalytic Codependencies of YAP/TAZ in Human Cancer
Abstract
The Hippo signaling pathway is commonly dysregulated in human cancer, which leads to a powerful tumor dependency on the YAP/TAZ transcriptional coactivators. In this study, we used paralog cotargeting CRISPR screens to identify kinases MARK2/3 as absolute catalytic requirements for YAP/TAZ function in diverse carcinoma and sarcoma contexts. Underlying this observation is the direct MARK2/3-dependent phosphorylation of NF2 and YAP/TAZ, which effectively reverses the tumor suppressive activity of the Hippo module kinases LATS1/2. To simulate targeting of MARK2/3, we adapted the CagA protein from Helicobacter pylori as a catalytic inhibitor of MARK2/3, which we show can regress established tumors in vivo. Together, these findings reveal MARK2/3 as powerful codependencies of YAP/TAZ in human cancer, targets that may allow for pharmacology that restores Hippo pathway-mediated tumor suppression. Significance: We show how genetic redundancy conceals tight functional relationships between signaling and transcriptional activation in cancer. Blocking the function of MARK2/3 kinases leads to the reactivation of the Hippo tumor suppressive pathway and may have therapeutic potential in YAP/TAZ-dysregulated carcinomas and sarcomas. See related commentary by Gauthier-Coles and Sheltzer, p. 2312.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
D.L. Spector reports grants from NIH during the conduct of the study. D.A. Tuveson reports other support from Leap Therapeutics, Xilis, Mestag Therapeutics, Dunad Therapeutics, and Sonata, as well as grants from ONO outside the submitted work. C.R. Vakoc reports grants from Treeline Biosciences during the conduct of the study, as well as personal fees from Treeline Biosciences and KSQ Therapeutics outside the submitted work. No disclosures were reported by the other authors.
Figures
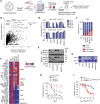



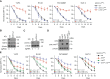

References
-
- Ma S, Meng Z, Chen R, Guan KL. The hippo pathway: biology and pathophysiology. Annu Rev Biochem 2019;88:577–604. - PubMed
-
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 2005;122:421–34. - PubMed
MeSH terms
Substances
Grants and funding
- R21 CA245859/CA/NCI NIH HHS/United States
- U01 CA224013/CA/NCI NIH HHS/United States
- R01 CA229699/CA/NCI NIH HHS/United States
- P30 CA045508/CA/NCI NIH HHS/United States
- U01 CA210240/CA/NCI NIH HHS/United States
- 2P01CA013106/National Cancer Institute (NCI)
- KL 3228/1-1/Deutsche Forschungsgemeinschaft (DFG)
- P30-CA045508/National Cancer Institute (NCI)
- CA45508/National Cancer Institute (NCI)
- R01 CA188134/CA/NCI NIH HHS/United States
- R01 CA281106/CA/NCI NIH HHS/United States
- R01 CA190092/CA/NCI NIH HHS/United States
- P01 CA013106/CA/NCI NIH HHS/United States
- CA013106/National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

