The chemokine Cxcl14 regulates interneuron differentiation in layer I of the somatosensory cortex
- PMID: 39058591
- PMCID: PMC11373301
- DOI: 10.1016/j.celrep.2024.114531
The chemokine Cxcl14 regulates interneuron differentiation in layer I of the somatosensory cortex
Abstract
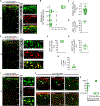
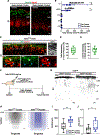
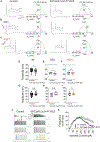
Spontaneous and sensory-evoked activity sculpts developing circuits. Yet, how these activity patterns intersect with cellular programs regulating the differentiation of neuronal subtypes is not well understood. Through electrophysiological and in vivo longitudinal analyses, we show that C-X-C motif chemokine ligand 14 (Cxcl14), a gene previously characterized for its association with tumor invasion, is expressed by single-bouquet cells (SBCs) in layer I (LI) of the somatosensory cortex during development. Sensory deprivation at neonatal stages markedly decreases Cxcl14 expression. Additionally, we report that loss of function of this gene leads to increased intrinsic excitability of SBCs-but not LI neurogliaform cells-and augments neuronal complexity. Furthermore, Cxcl14 loss impairs sensory map formation and compromises the in vivo recruitment of superficial interneurons by sensory inputs. These results indicate that Cxcl14 is required for LI differentiation and demonstrate the emergent role of chemokines as key players in cortical network development.
Keywords: CP: Neuroscience; barrel cortex; chemokines; development; interneurons; layer I; sensory inputs.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

