Nuclear pyruvate dehydrogenase complex regulates histone acetylation and transcriptional regulation in the ethylene response
- PMID: 39058774
- PMCID: PMC11277378
- DOI: 10.1126/sciadv.ado2825
Nuclear pyruvate dehydrogenase complex regulates histone acetylation and transcriptional regulation in the ethylene response
Abstract
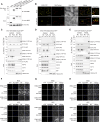
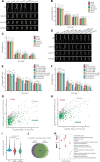
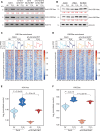
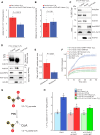
Ethylene plays its essential roles in plant development, growth, and defense responses by controlling the transcriptional reprograming, in which EIN2-C-directed regulation of histone acetylation is the first key step for chromatin to perceive ethylene signaling. But how the nuclear acetyl coenzyme A (acetyl CoA) is produced to ensure the ethylene-mediated histone acetylation is unknown. Here we report that ethylene triggers the accumulation of the pyruvate dehydrogenase complex (PDC) in the nucleus to synthesize nuclear acetyl CoA to regulate ethylene response. PDC is identified as an EIN2-C nuclear partner, and ethylene triggers its nuclear accumulation. Mutations in PDC lead to an ethylene hyposensitivity that results from the reduction of histone acetylation and transcription activation. Enzymatically active nuclear PDC synthesizes nuclear acetyl CoA for EIN2-C-directed histone acetylation and transcription regulation. These findings uncover a mechanism by which PDC-EIN2 converges the mitochondrial enzyme-mediated nuclear acetyl CoA synthesis with epigenetic and transcriptional regulation for plant hormone response.
Figures

Update of
-
Nuclear Pyruvate Dehydrogenase Complex Regulates Histone Acetylation and Transcriptional Regulation in the Ethylene Response.bioRxiv [Preprint]. 2024 May 6:2023.10.25.564010. doi: 10.1101/2023.10.25.564010. bioRxiv. 2024. Update in: Sci Adv. 2024 Jul 26;10(30):eado2825. doi: 10.1126/sciadv.ado2825. PMID: 37961310 Free PMC article. Updated. Preprint.
References
-
- Ecker J. R., The ethylene signal transduction pathway in plants. Science 268, 667–675 (1995). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

