Fetal hypoplastic lungs have multilineage inflammation that is reversed by amniotic fluid stem cell extracellular vesicle treatment
- PMID: 39058789
- PMCID: PMC11277482
- DOI: 10.1126/sciadv.adn5405
Fetal hypoplastic lungs have multilineage inflammation that is reversed by amniotic fluid stem cell extracellular vesicle treatment
Abstract
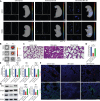
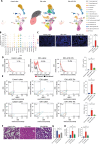
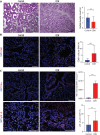
Antenatal administration of extracellular vesicles from amniotic fluid stem cells (AFSC-EVs) reverses features of pulmonary hypoplasia in models of congenital diaphragmatic hernia (CDH). However, it remains unknown which lung cellular compartments and biological pathways are affected by AFSC-EV therapy. Herein, we conducted single-nucleus RNA sequencing (snRNA-seq) on rat fetal CDH lungs treated with vehicle or AFSC-EVs. We identified that intra-amniotically injected AFSC-EVs reach the fetal lung in rats with CDH, where they promote lung branching morphogenesis and epithelial cell differentiation. Moreover, snRNA-seq revealed that rat fetal CDH lungs have a multilineage inflammatory signature with macrophage enrichment, which is reversed by AFSC-EV treatment. Macrophage enrichment in CDH fetal rat lungs was confirmed by immunofluorescence, flow cytometry, and inhibition studies with GW2580. Moreover, we validated macrophage enrichment in human fetal CDH lung autopsy samples. Together, this study advances knowledge on the pathogenesis of pulmonary hypoplasia and further evidence on the value of an EV-based therapy for CDH fetuses.
Figures



References
-
- Cotten C. M., Pulmonary hypoplasia. Semin. Fetal Neonatal Med. 22, 250–255 (2017). - PubMed
-
- Zani A., Chung W. K., Deprest J., Harting M. T., Jancelewicz T., Kunisaki S. M., Patel N., Antounians L., Puligandla P. S., Keijzer R., Congenital diaphragmatic hernia. Nat. Rev. Dis. Primers. 8, 37 (2022). - PubMed
-
- Jeanty C., Kunisaki S. M., MacKenzie T. C., Novel non-surgical prenatal approaches to treating congenital diaphragmatic hernia. Semin. Fetal Neonatal Med. 19, 349–356 (2014). - PubMed
-
- Figueira R. L., Antounians L., Zani-Ruttenstock E., Khalaj K., Zani A., Fetal lung regeneration using stem cell-derived extracellular vesicles: A new frontier for pulmonary hypoplasia secondary to congenital diaphragmatic hernia. Prenat. Diagn. 42, 364–372 (2022). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

