Update on gene fusions and the emerging clinicopathological landscape of peritoneal and pleural mesotheliomas and other neoplasms
- PMID: 39059063
- PMCID: PMC11326890
- DOI: 10.1016/j.esmoop.2024.103644
Update on gene fusions and the emerging clinicopathological landscape of peritoneal and pleural mesotheliomas and other neoplasms
Abstract
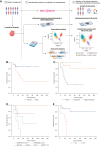
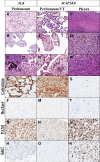
Background: Mesothelioma is a rare and aggressive malignant neoplasm arising from mesothelial cells, which occasionally manifests recurrent fusions. EWSR1/FUS-CREB, YY1, MAP3K8, NR4A3, and ALK-rearranged proliferations have been reported in limited series with no clear histological or clinical correlations, limiting clinicians' ability to assess prognosis and integrate these new entities into therapeutic decisions. The aim of this study was to better characterize these rearranged proliferations histologically, molecularly, and clinically.
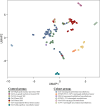
Methods: Clinical, pathological, and comprehensive transcriptome and mutation data were collected for each case.
Results: A total of 41 tumors were included, encompassing 7 ALK, 10 MAP3K8, 4 NR4A3, 8 ESWR1/FUS::ATF1, 8 EWSR1::YY1, and 4 SUFU-fused cases. We found a female predominance, except for cases harboring NR4A3 and SUFU; and most patients were around 60 years of age, but those harboring ALK or EWSR1/FUS::ATF1 gene fusions were younger. Each group exhibited distinct histological, immunohistochemical, molecular features, and oncological courses. Specifically, MAP3K8 and ALK presented PAX8+ papillary proliferations, ESWR1/FUS::ATF1 and EWSR1::YY1 displayed angiomatoid fibrous histiocytoma-like patterns, while SUFU showcased 'tissue culture'-like spindle cell proliferation. Poor prognosis factors were the pleural site, male sex, Ki67 ≥10%, and ESWR1/FUS::ATF1 or SUFU gene fusions.
Conclusions: This study significantly broadens the spectrum of mesothelial tumors associated with fusions, offering insight into novel epithelioid (mesothelial) proliferations with distinctive histological appearances, molecular profiles, and prognoses to guide adapted treatments for patients.
Keywords: ALK; EWSR1; MAP3K8; SUFU; gene fusion; mesothelioma.
Copyright © 2024 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Figures


References
-
- Robinson B.W.S., Lake R.A. Advances in malignant mesothelioma. N Engl J Med. 2005;353(15):1591–1603. - PubMed
-
- Tsao A.S., Lindwasser O.W., Adjei A.A., et al. Current and future management of malignant mesothelioma: a consensus report from the National Cancer Institute Thoracic Malignancy Steering Committee, International Association for the Study of Lung Cancer, and Mesothelioma Applied Research Foundation. J Thorac Oncol. 2018;13(11):1655–1667. - PubMed
-
- Opitz I., Bille A., Dafni U., et al. European epidemiology of pleural mesothelioma—real-life data from a Joint Analysis of the Mesoscape Database of the European Thoracic Oncology Platform and the European Society of Thoracic Surgery Mesothelioma Database. J Thorac Oncol. 2023;18(9):1233–1247. - PubMed
-
- Bueno R., Stawiski E.W., Goldstein L.D., et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48(4):407–416. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

