Human microglial cells as a therapeutic target in a neurodevelopmental disease model
- PMID: 39059378
- PMCID: PMC11368698
- DOI: 10.1016/j.stemcr.2024.06.013
Human microglial cells as a therapeutic target in a neurodevelopmental disease model
Abstract
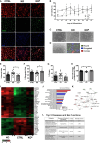
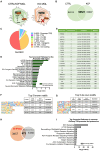
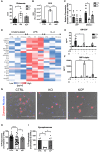
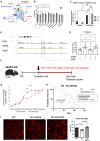
Although microglia are macrophages of the central nervous system, their involvement is not limited to immune functions. The roles of microglia during development in humans remain poorly understood due to limited access to fetal tissue. To understand how microglia can impact human neurodevelopment, the methyl-CpG binding protein 2 (MECP2) gene was knocked out in human microglia-like cells (MGLs). Disruption of the MECP2 in MGLs led to transcriptional and functional perturbations, including impaired phagocytosis. The co-culture of healthy MGLs with MECP2-knockout (KO) neurons rescued synaptogenesis defects, suggesting a microglial role in synapse formation. A targeted drug screening identified ADH-503, a CD11b agonist, restored phagocytosis and synapse formation in spheroid-MGL co-cultures, significantly improved disease progression, and increased survival in MeCP2-null mice. These results unveil a MECP2-specific regulation of human microglial phagocytosis and identify a novel therapeutic treatment for MECP2-related conditions.
Keywords: ADH-503; CD11b; MECP2; chromatin; iPSC; integrin; microglia; neurodevelopment; neurons; phagocytosis; stem cells; synaptogenesis.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.R.M. is a co-founder and has an equity interest in TISMOO, a company dedicated to genetic analysis and human brain organogenesis focusing on therapeutic applications customized for the disorder autism spectrum and other neurological disorders origin genetics. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict-of-interest policies. The authors have a patent application in works related to this publication.
Figures

References
-
- Alliot F., Godin I., Pessac B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res. Dev. Brain Res. 1999;117:145–152. http://www.ncbi.nlm.nih.gov/pubmed/10567732 - PubMed
-
- Butovsky O., Jedrychowski M.P., Moore C.S., Cialic R., Lanser A.J., Gabriely G., Koeglsperger T., Dake B., Wu P.M., Doykan C.E., et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

