Natural malaria infection elicits rare but potent neutralizing antibodies to the blood-stage antigen RH5
- PMID: 39059381
- PMCID: PMC11383431
- DOI: 10.1016/j.cell.2024.06.037
Natural malaria infection elicits rare but potent neutralizing antibodies to the blood-stage antigen RH5
Abstract
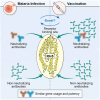
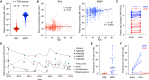
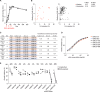
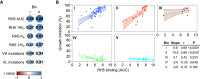
Plasmodium falciparum reticulocyte-binding protein homolog 5 (RH5) is the most advanced blood-stage malaria vaccine candidate and is being evaluated for efficacy in endemic regions, emphasizing the need to study the underlying antibody response to RH5 during natural infection, which could augment or counteract responses to vaccination. Here, we found that RH5-reactive B cells were rare, and circulating immunoglobulin G (IgG) responses to RH5 were short-lived in malaria-exposed Malian individuals, despite repeated infections over multiple years. RH5-specific monoclonal antibodies isolated from eight malaria-exposed individuals mostly targeted non-neutralizing epitopes, in contrast to antibodies isolated from five RH5-vaccinated, malaria-naive UK individuals. However, MAD8-151 and MAD8-502, isolated from two malaria-exposed Malian individuals, were among the most potent neutralizers out of 186 antibodies from both cohorts and targeted the same epitopes as the most potent vaccine-induced antibodies. These results suggest that natural malaria infection may boost RH5-vaccine-induced responses and provide a clear strategy for the development of next-generation RH5 vaccines.
Keywords: Plasmodium falciparum; RH5; malaria; monoclonal antibodies; natural infection; vaccine design.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests J.T., L.T.W., and A.J.R.C. are co-inventors on a provisional patent filed on the mAbs described in this study. J.R.B., K.M., M.K.H., and S.J.D. are inventors on patent applications relating to RH5 malaria vaccines and/or antibodies. A.M.M. has an immediate family member who is an inventor on patent applications relating to RH5 malaria vaccines and antibodies. The content of this publication does not necessarily reflect the views or policies of the DHHS or of the institutions and companies with which the authors are affiliated.
Figures







References
-
- World Health Organization . 2023. World malaria report 2023.
-
- Datoo M.S., Natama H.M., Somé A., Bellamy D., Traoré O., Rouamba T., Tahita M.C., Ido N.F.A., Yameogo P., Valia D., et al. Efficacy and immunogenicity of R21/Matrix-M vaccine against clinical malaria after 2 years' follow-up in children in Burkina Faso: a phase 1/2b randomised controlled trial. Lancet Infect. Dis. 2022;22:1728–1736. doi: 10.1016/S1473-3099(22)00442-X. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

