Longitudinal integrative cell-free DNA analysis in gestational diabetes mellitus
- PMID: 39059385
- PMCID: PMC11384941
- DOI: 10.1016/j.xcrm.2024.101660
Longitudinal integrative cell-free DNA analysis in gestational diabetes mellitus
Abstract
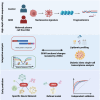
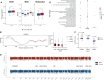
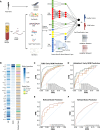
Gestational diabetes mellitus (GDM) presents varied manifestations throughout pregnancy and poses a complex clinical challenge. High-depth cell-free DNA (cfDNA) sequencing analysis holds promise in advancing our understanding of GDM pathogenesis and prediction. In 299 women with GDM and 299 matched healthy pregnant women, distinct cfDNA fragment characteristics associated with GDM are identified throughout pregnancy. Integrating cfDNA profiles with lipidomic and single-cell transcriptomic data elucidates functional changes linked to altered lipid metabolism processes in GDM. Transcription start site (TSS) scores in 50 feature genes are used as the cfDNA signature to distinguish GDM cases from controls effectively. Notably, differential coverage of the islet acinar marker gene PRSS1 emerges as a valuable biomarker for GDM. A specialized neural network model is developed, predicting GDM occurrence and validated across two independent cohorts. This research underscores the high-depth cfDNA early prediction and characterization of GDM, offering insights into its molecular underpinnings and potential clinical applications.
Keywords: PRSS1; cell-free DNA; gestational diabetes mellitus; pregnancy dynamics.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Federation I. tenth Edition. International Diabetes; 2021. IDF Diabetes Atlas.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

