Discovery and validation of a 10-gene predictive signature for response to adjuvant chemotherapy in stage II and III colon cancer
- PMID: 39059386
- PMCID: PMC11384724
- DOI: 10.1016/j.xcrm.2024.101661
Discovery and validation of a 10-gene predictive signature for response to adjuvant chemotherapy in stage II and III colon cancer
Abstract
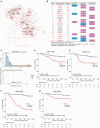
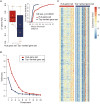
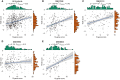
Identifying patients with stage II and III colon cancer who will benefit from 5-fluorouracil (5-FU)-based adjuvant chemotherapy is crucial for the advancement of personalized cancer therapy. We employ a semi-supervised machine learning approach to analyze a large dataset with 933 stage II and III colon cancer samples. Our analysis leverages gene regulatory networks to discover an 18-gene prognostic signature and to explore a 10-gene signature that potentially predicts chemotherapy benefits. The 10-gene signature demonstrates strong prognostic power and shows promising potential to predict chemotherapy benefits. We establish a robust clinical assay on the NanoString nCounter platform, validated in a retrospective formalin-fixed paraffin-embedded (FFPE) cohort, which represents an important step toward clinical application. Our study lays the groundwork for improving adjuvant chemotherapy and potentially expanding into immunotherapy decision-making in colon cancer. Future prospective studies are needed to validate and establish the clinical utility of the 10-gene signature in clinical settings.
Keywords: adjuvant therapy; biomarkers; chemotherapy; colon cancer; gene expression; immune checkpoint blockade; machine learning; prediction; prognosis.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Meyerhardt J.A., Shi Q., Fuchs C.S., Meyer J., Niedzwiecki D., Zemla T., Kumthekar P., Guthrie K.A., Couture F., Kuebler P., et al. Effect of Celecoxib vs Placebo Added to Standard Adjuvant Therapy on Disease-Free Survival Among Patients With Stage III Colon Cancer: The CALGB/SWOG 80702 (Alliance) Randomized Clinical Trial. JAMA. 2021;325:1277–1286. doi: 10.1001/jama.2021.2454. - DOI - PMC - PubMed
-
- Iveson T.J., Sobrero A.F., Yoshino T., Souglakos I., Ou F.S., Meyers J.P., Shi Q., Grothey A., Saunders M.P., Labianca R., et al. Duration of Adjuvant Doublet Chemotherapy (3 or 6 months) in Patients With High-Risk Stage II Colorectal Cancer. J. Clin. Oncol. 2021;39:631–641. doi: 10.1200/JCO.20.01330. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

