Regulation of Caenorhabditis elegans HLH-30 subcellular localization dynamics: Evidence for a redox-dependent mechanism
- PMID: 39059513
- PMCID: PMC11977398
- DOI: 10.1016/j.freeradbiomed.2024.07.027
Regulation of Caenorhabditis elegans HLH-30 subcellular localization dynamics: Evidence for a redox-dependent mechanism
Abstract
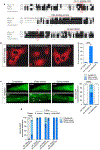
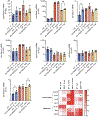
Basic Helix-Loop-Helix (bHLH) transcription factors TFEB/TFE3 and HLH-30 are key regulators of autophagy induction and lysosomal biogenesis in mammals and C. elegans, respectively. While much is known about the regulation of TFEB/TFE3, how HLH-30 subcellular dynamics and transactivation are modulated are yet poorly understood. Thus, elucidating the regulation of C. elegans HLH-30 will provide evolutionary insight into the mechanisms governing the function of bHLH transcription factor family. We report here that HLH-30 is retained in the cytoplasm mainly through its conserved Ser201 residue and that HLH-30 physically interacts with the 14-3-3 protein FTT-2 in this location. The FoxO transcription factor DAF-16 is not required for HLH-30 nuclear translocation upon stress, despite that both proteins partner to form a complex that coordinately regulates several organismal responses. Similar as described for DAF-16, the importin IMB-2 assists HLH-30 nuclear translocation, but constitutive HLH-30 nuclear localization is not sufficient to trigger its distinctive transcriptional response. Furthermore, we identify FTT-2 as the target of diethyl maleate (DEM), a GSH depletor that causes a transient nuclear translocation of HLH-30. Together, our work demonstrates that the regulation of TFEB/TFE3 and HLH-30 family members is evolutionarily conserved and that, in addition to a direct redox regulation through its conserved single cysteine residue, HLH-30 can also be indirectly regulated by a redox-dependent mechanism, probably through FTT-2 oxidation.
Keywords: 14-3-3 proteins; DAF-16; Diethyl maleate; HLH-30; Redox.
Copyright © 2024. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest Authors declare no competing interests
Figures


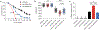


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

