Gene expression and chromatin conformation of microglia in virally suppressed people with HIV
- PMID: 39060113
- PMCID: PMC11282357
- DOI: 10.26508/lsa.202402736
Gene expression and chromatin conformation of microglia in virally suppressed people with HIV
Erratum in
-
Correction: Gene expression and chromatin conformation of microglia in virally suppressed people with HIV.Life Sci Alliance. 2024 Oct 28;8(1):e202403085. doi: 10.26508/lsa.202403085. Print 2025 Jan. Life Sci Alliance. 2024. PMID: 39467638 Free PMC article.
Abstract
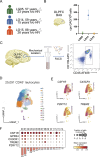
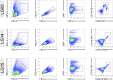
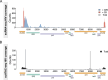
The presence of HIV in sequestered reservoirs is a central impediment to a functional cure, allowing HIV to persist despite life-long antiretroviral therapy (ART), and driving a variety of comorbid conditions. Our understanding of the latent HIV reservoir in the central nervous system is incomplete, because of difficulties in accessing human central nervous system tissues. Microglia contribute to HIV reservoirs, but the molecular phenotype of HIV-infected microglia is poorly understood. We leveraged the unique "Last Gift" rapid autopsy program, in which people with HIV are closely followed until days or even hours before death. Microglial populations were heterogeneous regarding their gene expression profiles but showed similar chromatin accessibility landscapes. Despite ART, we detected occasional microglia containing cell-associated HIV RNA and HIV DNA integrated into open regions of the host's genome (∼0.005%). Microglia with detectable HIV RNA showed an inflammatory phenotype. These results demonstrate a distinct myeloid cell reservoir in the brains of people with HIV despite suppressive ART. Strategies for curing HIV and neurocognitive impairment will need to consider the myeloid compartment to be successful.
© 2024 Schlachetzki et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Angelovich TA, Cochrane CR, Zhou J, Tumpach C, Byrnes SJ, Jamal Eddine J, Waring E, Busman-Sahay K, Deleage C, Jenkins TA, et al. (2023) Regional analysis of intact and defective HIV proviruses in the brain of viremic and virally suppressed people with HIV. Ann Neurol 94: 798–802. 10.1002/ana.26750 - DOI - PMC - PubMed
-
- Churchill MJ, Gorry PR, Cowley D, Lal L, Sonza S, Purcell DF, Thompson KA, Gabuzda D, McArthur JC, Pardo CA, et al. (2006) Use of laser capture microdissection to detect integrated HIV-1 DNA in macrophages and astrocytes from autopsy brain tissues. J Neurovirol 12: 146–152. 10.1080/13550280600748946 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- UM1 AI164559/AI/NIAID NIH HHS/United States
- U24 MH100928/MH/NIMH NIH HHS/United States
- P01 AI169609/AI/NIAID NIH HHS/United States
- RF1 AG061060/AG/NIA NIH HHS/United States
- P30 MH062512/MH/NIMH NIH HHS/United States
- P30 AI036214/AI/NIAID NIH HHS/United States
- R01 DA055491/DA/NIDA NIH HHS/United States
- R01 DA057337/DA/NIDA NIH HHS/United States
- UL1 TR001442/TR/NCATS NIH HHS/United States
- K99 HD111686/HD/NICHD NIH HHS/United States
- R61 DA058501/DA/NIDA NIH HHS/United States
- R21 MH134401/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
