Single cell transcriptomic profiling identifies tumor-acquired and therapy-resistant cell states in pediatric rhabdomyosarcoma
- PMID: 39060228
- PMCID: PMC11282092
- DOI: 10.1038/s41467-024-50527-2
Single cell transcriptomic profiling identifies tumor-acquired and therapy-resistant cell states in pediatric rhabdomyosarcoma
Abstract
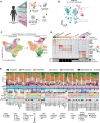
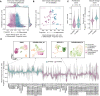
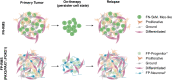
Rhabdomyosarcoma (RMS) is a pediatric tumor that resembles undifferentiated muscle cells; yet the extent to which cell state heterogeneity is shared with human development has not been described. Using single-cell/nucleus RNA sequencing from patient tumors, patient-derived xenografts, primary in vitro cultures, and cell lines, we identify four dominant muscle-lineage cell states: progenitor, proliferative, differentiated, and ground cells. We stratify these RMS cells/nuclei along the continuum of human muscle development and show that they share expression patterns with fetal/embryonal myogenic precursors rather than postnatal satellite cells. Fusion-negative RMS (FN-RMS) have a discrete stem cell hierarchy that recapitulates fetal muscle development and contain therapy-resistant FN-RMS progenitors that share transcriptomic similarity with bipotent skeletal mesenchymal cells. Fusion-positive RMS have tumor-acquired cells states, including a neuronal cell state, that are not found in myogenic development. This work identifies previously underappreciated cell state heterogeneity including unique treatment-resistant and tumor-acquired cell states that differ across RMS subtypes.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Grants and funding
- 2022 SFA 13-22/Sarcoma Foundation of America (Sarcoma Foundation of America, Inc.)
- DRSG-33P-20/Damon Runyon Cancer Research Foundation (Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation)
- R01CA269213, R01CA276116, U54CA231630/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 CA269213/CA/NCI NIH HHS/United States
- K99CA278696/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

