A TNIP1-driven systemic autoimmune disorder with elevated IgG4
- PMID: 39060650
- PMCID: PMC11362012
- DOI: 10.1038/s41590-024-01902-0
A TNIP1-driven systemic autoimmune disorder with elevated IgG4
Abstract
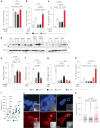
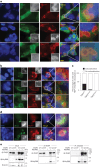
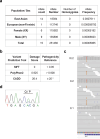
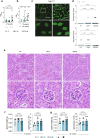
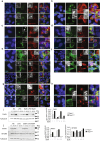
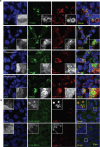
Whole-exome sequencing of two unrelated kindreds with systemic autoimmune disease featuring antinuclear antibodies with IgG4 elevation uncovered an identical ultrarare heterozygous TNIP1Q333P variant segregating with disease. Mice with the orthologous Q346P variant developed antinuclear autoantibodies, salivary gland inflammation, elevated IgG2c, spontaneous germinal centers and expansion of age-associated B cells, plasma cells and follicular and extrafollicular helper T cells. B cell phenotypes were cell-autonomous and rescued by ablation of Toll-like receptor 7 (TLR7) or MyD88. The variant increased interferon-β without altering nuclear factor kappa-light-chain-enhancer of activated B cells signaling, and impaired MyD88 and IRAK1 recruitment to autophagosomes. Additionally, the Q333P variant impaired TNIP1 localization to damaged mitochondria and mitophagosome formation. Damaged mitochondria were abundant in the salivary epithelial cells of Tnip1Q346P mice. These findings suggest that TNIP1-mediated autoimmunity may be a consequence of increased TLR7 signaling due to impaired recruitment of downstream signaling molecules and damaged mitochondria to autophagosomes and may thus respond to TLR7-targeted therapeutics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

