An intestinal TH17 cell-derived subset can initiate cancer
- PMID: 39060651
- PMCID: PMC11362008
- DOI: 10.1038/s41590-024-01909-7
An intestinal TH17 cell-derived subset can initiate cancer
Abstract
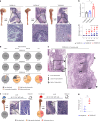
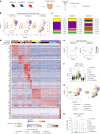
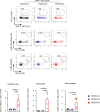
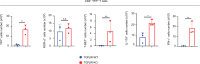
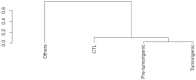
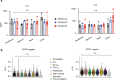
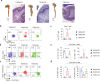
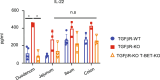
Approximately 25% of cancers are preceded by chronic inflammation that occurs at the site of tumor development. However, whether this multifactorial oncogenic process, which commonly occurs in the intestines, can be initiated by a specific immune cell population is unclear. Here, we show that an intestinal T cell subset, derived from interleukin-17 (IL-17)-producing helper T (TH17) cells, induces the spontaneous transformation of the intestinal epithelium. This subset produces inflammatory cytokines, and its tumorigenic potential is not dependent on IL-17 production but on the transcription factors KLF6 and T-BET and interferon-γ. The development of this cell type is inhibited by transforming growth factor-β1 (TGFβ1) produced by intestinal epithelial cells. TGFβ signaling acts on the pretumorigenic TH17 cell subset, preventing its progression to the tumorigenic stage by inhibiting KLF6-dependent T-BET expression. This study therefore identifies an intestinal T cell subset initiating cancer.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

