Fibroblasts Promote Resistance to KRAS Silencing in Colorectal Cancer Cells
- PMID: 39061234
- PMCID: PMC11274566
- DOI: 10.3390/cancers16142595
Fibroblasts Promote Resistance to KRAS Silencing in Colorectal Cancer Cells
Abstract
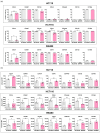
Colorectal cancer (CRC) responses to KRAS-targeted inhibition have been limited due to low response rates, the mechanisms of which remain unknown. Herein, we explored the cancer-associated fibroblasts (CAFs) secretome as a mediator of resistance to KRAS silencing. CRC cell lines HCT15, HCT116, and SW480 were cultured either in recommended media or in conditioned media from a normal colon fibroblast cell line (CCD-18Co) activated with rhTGF-β1 to induce a CAF-like phenotype. The expression of membrane stem cell markers was analyzed by flow cytometry. Stem cell potential was evaluated by a sphere formation assay. RNAseq was performed in KRAS-silenced HCT116 colonospheres treated with either control media or conditioned media from CAFs. Our results demonstrated that KRAS-silencing up-regulated CD24 and down-regulated CD49f and CD104 in the three cell lines, leading to a reduction in sphere-forming efficiency. However, CAF-secreted factors restored stem cell marker expression and increased stemness. RNA sequencing showed that CAF-secreted factors up-regulated genes associated with pro-tumorigenic pathways in KRAS-silenced cells, including KRAS, TGFβ, NOTCH, WNT, MYC, cell cycle progression and exit from quiescence, epithelial-mesenchymal transition, and immune regulation. Overall, our results suggest that resistance to KRAS-targeted inhibition might derive not only from cell-intrinsic causes but also from external elements, such as fibroblast-secreted factors.
Keywords: KRAS; cancer stemness; colorectal cancer; epithelial-mesenchymal transition; fibroblasts; therapy resistance.
Conflict of interest statement
The authors declare no conflict of interest. A.S.R. contributed to this work while previously affiliated with i3S and IPATIMUP. A.S.R. is currently affiliated with and employed by Novartis. Novartis did not fund or contribute to this work.
Figures





References
-
- Patricelli M.P., Janes M.R., Li L.S., Hansen R., Peters U., Kessler L.V., Chen Y., Kucharski J.M., Feng J., Ely T., et al. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discov. 2016;6:316–329. doi: 10.1158/2159-8290.CD-15-1105. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

