The Surviving, Not Thriving, Photoreceptors in Patients with ABCA4 Stargardt Disease
- PMID: 39061682
- PMCID: PMC11275370
- DOI: 10.3390/diagnostics14141545
The Surviving, Not Thriving, Photoreceptors in Patients with ABCA4 Stargardt Disease
Abstract
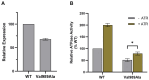
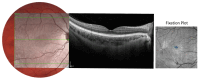
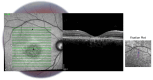
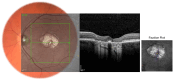
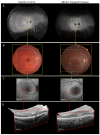
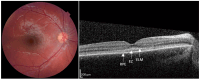
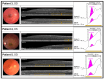
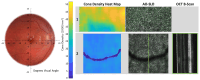
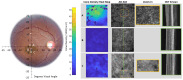
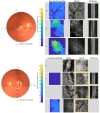
Stargardt disease (STGD1), associated with biallelic variants in the ABCA4 gene, is the most common heritable macular dystrophy and is currently untreatable. To identify potential treatment targets, we characterized surviving STGD1 photoreceptors. We used clinical data to identify macular regions with surviving STGD1 photoreceptors. We compared the hyperreflective bands in the optical coherence tomographic (OCT) images that correspond to structures in the STGD1 photoreceptor inner segments to those in controls. We used adaptive optics scanning light ophthalmoscopy (AO-SLO) to study the distribution of cones and AO-OCT to evaluate the interface of photoreceptors and retinal pigment epithelium (RPE). We found that the profile of the hyperreflective bands differed dramatically between patients with STGD1 and controls. AO-SLOs showed patches in which cone densities were similar to those in healthy retinas and others in which the cone population was sparse. In regions replete with cones, there was no debris at the photoreceptor-RPE interface. In regions with sparse cones, there was abundant debris. Our results raise the possibility that pharmaceutical means may protect surviving photoreceptors and so mitigate vision loss in patients with STGD1.
Keywords: Stargardt disease; inherited retinal disease; mitochondria; optical coherence tomography adaptive optics; photoreceptors; retina; retinal imaging.
Conflict of interest statement
M.M. (Mircea Mujat): Physical Sciences Inc. (PSI) (E, P). All other authors: no conflicts of interest.
Figures

References
-
- Li C.H.Z., Pas J.A.A.H., Corradi Z., Hitti-Malin R.J., Hoogstede A., Runhart E.H., Dhooge P.P.A., Collin R.W.J., Cremers F.P.M., Hoyng C.B. Study of Late-Onset Stargardt Type 1 Disease: Characteristics, Genetics, and Progression. Ophthalmology. 2024;131:87–97. doi: 10.1016/j.ophtha.2023.08.011. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

