The Association of Neonatal Gut Microbiota Community State Types with Birth Weight
- PMID: 39062221
- PMCID: PMC11276374
- DOI: 10.3390/children11070770
The Association of Neonatal Gut Microbiota Community State Types with Birth Weight
Abstract
Background: while most gut microbiota research has focused on term infants, the health outcomes of preterm infants are equally important. Very-low-birth-weight (VLBW) or extremely-low-birth-weight (ELBW) preterm infants have a unique gut microbiota structure, and probiotics have been reported to somewhat accelerate the maturation of the gut microbiota and reduce intestinal inflammation in very-low preterm infants, thereby improving their long-term outcomes. The aim of this study was to investigate the structure of gut microbiota in ELBW neonates to facilitate the early identification of different types of low-birth-weight (LBW) preterm infants.
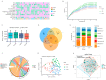
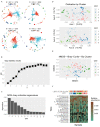
Methods: a total of 98 fecal samples from 39 low-birth-weight preterm infants were included in this study. Three groups were categorized according to different birth weights: ELBW (n = 39), VLBW (n = 39), and LBW (n = 20). The gut microbiota structure of neonates was obtained by 16S rRNA gene sequencing, and microbiome analysis was conducted. The community state type (CST) of the microbiota was predicted, and correlation analysis was conducted with clinical indicators. Differences in the gut microbiota composition among ELBW, VLBW, and LBW were compared. The value of gut microbiota composition in the diagnosis of extremely low birth weight was assessed via a random forest-machine learning approach.
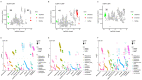
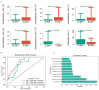
Results: we briefly analyzed the structure of the gut microbiota of preterm infants with low birth weight and found that the ELBW, VLBW, and LBW groups exhibited gut microbiota with heterogeneous compositions. Low-birth-weight preterm infants showed five CSTs dominated by Enterococcus, Staphylococcus, Klebsiella, Streptococcus, Pseudescherichia, and Acinetobacter. The birth weight and clinical indicators related to prematurity were associated with the CST. We found the composition of the gut microbiota was specific to the different types of low-birth-weight premature infants, namely, ELBW, VLBW, and LBW. The ELBW group exhibited significantly more of the potentially harmful intestinal bacteria Acinetobacter relative to the VLBW and LBW groups, as well as a significantly lower abundance of the intestinal probiotic Bifidobacterium. Based on the gut microbiota's composition and its correlation with low weight, we constructed random forest model classifiers to distinguish ELBW and VLBW/LBW infants. The area under the curve of the classifiers constructed with Enterococcus, Klebsiella, and Acinetobacter was found to reach 0.836 by machine learning evaluation, suggesting that gut microbiota composition may be a potential biomarker for ELBW preterm infants.
Conclusions: the gut bacteria of preterm infants showed a CST with Enterococcus, Klebsiella, and Acinetobacter as the dominant genera. ELBW preterm infants exhibit an increase in the abundance of potentially harmful bacteria in the gut and a decrease in beneficial bacteria. These potentially harmful bacteria may be potential biomarkers for ELBW preterm infants.
Keywords: 16S rRNA gene sequencing; community state type; gut microbiota; infant; low birth weight; machine learning; neonate.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Dynamics alteration of the gut microbiota and faecal metabolomes in very low or extremely low birth weight infants: a Chinese single-center, prospective cohort study.Front Microbiol. 2024 Aug 23;15:1438213. doi: 10.3389/fmicb.2024.1438213. eCollection 2024. Front Microbiol. 2024. PMID: 39247697 Free PMC article.
-
Multi-omics analyses of gut microbiota via 16S rRNA gene sequencing, LC-MS/MS and diffusion tension imaging reveal aberrant microbiota-gut-brain axis in very low or extremely low birth weight infants with white matter injury.BMC Microbiol. 2023 Dec 6;23(1):387. doi: 10.1186/s12866-023-03103-5. BMC Microbiol. 2023. PMID: 38057706 Free PMC article.
-
Early gut microbiota in very low and extremely low birth weight preterm infants with feeding intolerance: a prospective case-control study.J Microbiol. 2022 Oct;60(10):1021-1031. doi: 10.1007/s12275-022-2180-2. Epub 2022 Aug 19. J Microbiol. 2022. PMID: 35984614 Free PMC article.
-
Establishment of an ideal gut microbiota to boost healthy growth of neonates.Crit Rev Microbiol. 2019 Feb;45(1):118-129. doi: 10.1080/1040841X.2018.1561643. Epub 2019 Feb 18. Crit Rev Microbiol. 2019. PMID: 30773108 Review.
-
Assessing the effect of disease on nutrition of the preterm infant.Clin Biochem. 1996 Oct;29(5):399-417. doi: 10.1016/0009-9120(96)00062-8. Clin Biochem. 1996. PMID: 8884061 Review.
Cited by
-
Uncovering the Effects of Different Formulae of Milk Powders on the Fecal Microorganisms and Metabolites of Bengal Tiger (Panthera tigris spp. tigris) Cubs.Animals (Basel). 2025 Apr 4;15(7):1053. doi: 10.3390/ani15071053. Animals (Basel). 2025. PMID: 40218446 Free PMC article.
References
-
- Pammi M., Cope J., Tarr P.I., Warner B.B., Morrow A.L., Mai V., Gregory K.E., Kroll J.S., McMurtry V., Ferris M.J., et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: A systematic review and meta-analysis. Microbiome. 2017;5:31. doi: 10.1186/s40168-017-0248-8. - DOI - PMC - PubMed
-
- Stewart C.J., Embleton N.D., Marrs E.C.L., Smith D.P., Fofanova T., Nelson A., Skeath T., Perry J.D., Petrosino J.F., Berrington J.E., et al. Longitudinal development of the gut microbiome and metabolome in preterm neonates with late onset sepsis and healthy controls. Microbiome. 2017;5:75. doi: 10.1186/s40168-017-0295-1. - DOI - PMC - PubMed
-
- Norman M., Hallberg B., Abrahamsson T., Björklund L.J., Domellöf M., Farooqi A., Foyn Bruun C., Gadsbøll C., Hellström-Westas L., Ingemansson F., et al. Association Between Year of Birth and 1-Year Survival Among Extremely Preterm Infants in Sweden During 2004–2007 and 2014–2016. JAMA. 2019;321:1188–1199. doi: 10.1001/jama.2019.2021. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

