In Silico Comparative Analysis of Ivermectin and Nirmatrelvir Inhibitors Interacting with the SARS-CoV-2 Main Protease
- PMID: 39062468
- PMCID: PMC11274663
- DOI: 10.3390/biom14070755
In Silico Comparative Analysis of Ivermectin and Nirmatrelvir Inhibitors Interacting with the SARS-CoV-2 Main Protease
Abstract
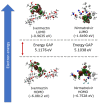
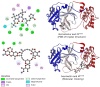
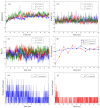
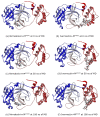
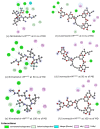
Exploring therapeutic options is crucial in the ongoing COVID-19 pandemic caused by SARS-CoV-2. Nirmatrelvir, which is a potent inhibitor that targets the SARS-CoV-2 Mpro, shows promise as an antiviral treatment. Additionally, Ivermectin, which is a broad-spectrum antiparasitic drug, has demonstrated effectiveness against the virus in laboratory settings. However, its clinical implications are still debated. Using computational methods, such as molecular docking and 100 ns molecular dynamics simulations, we investigated how Nirmatrelvir and Ivermectin interacted with SARS-CoV-2 Mpro(A). Calculations using density functional theory were instrumental in elucidating the behavior of isolated molecules, primarily by analyzing the frontier molecular orbitals. Our analysis revealed distinct binding patterns: Nirmatrelvir formed strong interactions with amino acids, like MET49, MET165, HIS41, HIS163, HIS164, PHE140, CYS145, GLU166, and ASN142, showing stable binding, with a root-mean-square deviation (RMSD) of around 2.0 Å. On the other hand, Ivermectin interacted with THR237, THR239, LEU271, LEU272, and LEU287, displaying an RMSD of 1.87 Å, indicating enduring interactions. Both ligands stabilized Mpro(A), with Ivermectin showing stability and persistent interactions despite forming fewer hydrogen bonds. These findings offer detailed insights into how Nirmatrelvir and Ivermectin bind to the SARS-CoV-2 main protease, providing valuable information for potential therapeutic strategies against COVID-19.
Keywords: Ivermectin; Nirmatrelvir; SARS-CoV-2; main protease (Mpro); molecular docking; molecular dynamics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Louten J. Essential Human Virology. Academic Press; Cambridge, MA, USA: 2016. Virus transmission and epidemiology; pp. 71–92. - DOI
-
- Asadi S. Airborne Infectious Disease Transmission via Expiratory Aerosol Particles and Aerosolized Fomites. University of California; Davis, CA, USA: 2020.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

