Effect of Health Status and Heat-Induced Inactivation on the Proteomic Profile of Plasma Rich in Growth Factors Obtained from Donors with Chronic Inflammatory Skin Conditions
- PMID: 39062477
- PMCID: PMC11275043
- DOI: 10.3390/biom14070763
Effect of Health Status and Heat-Induced Inactivation on the Proteomic Profile of Plasma Rich in Growth Factors Obtained from Donors with Chronic Inflammatory Skin Conditions
Abstract
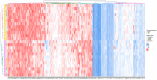
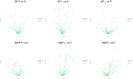
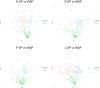
Atopic dermatitis, psoriasis and lichen sclerosus are among the most challenging conditions treated by dermatologists worldwide, with potentially significant physical, social and psychological impacts. Emerging evidence suggests that autologous-platelet-rich plasma could be used to manage skin inflammation. However, the presence of soluble autoimmune components could hinder their therapeutic potential. The aim of this study was to analyze the proteomic profile of plasma rich in growth factors (PRGFs) obtained from donors with inflammatory skin conditions to evaluate the impact of skin health status on the composition and bioactivity of PRGF-based treatments. Venous blood from healthy volunteers and patients with psoriasis, lichen sclerosus and atopic dermatitis was processed to produce PRGF supernatant. Half of the samples were subjected to an additional thermal treatment (56 °C) to inactivate inflammatory and immune molecules. Proteomic analysis was performed to assess the protein profile of PRGFs from healthy and non-healthy patients and the effect of Immunosafe treatment. Differential abundance patterns of several proteins related to key biological processes have been identified, including complement activation, blood coagulation, and glycolysis- and gluconeogenesis-related genes. These results also demonstrate that the thermal treatment (Immunosafe) contributes to the inactivation of the complement system and, as a consequence, reduction in the immunogenic potential of PRGF products.
Keywords: atopic dermatitis; lichen sclerosus; platelet-rich plasma; proteome; psoriasis; skin inflammation; thermal inactivation.
Conflict of interest statement
E.A. is the scientific director of the BTI Biotechnology Institute, the company that developed Endoret®PRGF® technology. R.T. and M.H.A. are researchers at BTI.
Figures




Similar articles
-
Bioactive Effect of Plasma-Rich in Growth Factors (PRGFs) on Cell-Based In Vitro Models of Skin Inflammation in Relation to Inflammatory Skin Disorders.Cureus. 2024 Nov 22;16(11):e74252. doi: 10.7759/cureus.74252. eCollection 2024 Nov. Cureus. 2024. PMID: 39712761 Free PMC article.
-
Impact of inflammatory skin conditions on the biological profile of plasma rich in growth factor.Tissue Cell. 2024 Dec;91:102560. doi: 10.1016/j.tice.2024.102560. Epub 2024 Sep 14. Tissue Cell. 2024. PMID: 39299031
-
Effects of heat-treatment on plasma rich in growth factors-derived autologous eye drop.Exp Eye Res. 2014 Feb;119:27-34. doi: 10.1016/j.exer.2013.12.005. Epub 2013 Dec 15. Exp Eye Res. 2014. PMID: 24345372
-
Progress in the use of plasma rich in growth factors in ophthalmology: from ocular surface to ocular fundus.Expert Opin Biol Ther. 2022 Jan;22(1):31-45. doi: 10.1080/14712598.2021.1945030. Epub 2021 Jul 19. Expert Opin Biol Ther. 2022. PMID: 34275392 Review.
-
PRGF exerts more potent proliferative and anti-inflammatory effects than autologous serum on a cell culture inflammatory model.Exp Eye Res. 2016 Oct;151:115-21. doi: 10.1016/j.exer.2016.08.012. Epub 2016 Aug 25. Exp Eye Res. 2016. PMID: 27567559 Review.
Cited by
-
The Role of Platelet Concentrates and Growth Factors in Facial Rejuvenation: A Systematic Review with Case Series.Medicina (Kaunas). 2025 Jan 7;61(1):84. doi: 10.3390/medicina61010084. Medicina (Kaunas). 2025. PMID: 39859067 Free PMC article.
-
Bioactive Effect of Plasma-Rich in Growth Factors (PRGFs) on Cell-Based In Vitro Models of Skin Inflammation in Relation to Inflammatory Skin Disorders.Cureus. 2024 Nov 22;16(11):e74252. doi: 10.7759/cureus.74252. eCollection 2024 Nov. Cureus. 2024. PMID: 39712761 Free PMC article.
References
-
- Liu Y., Wang H., Taylor M., Cook C., Martínez-Berdeja A., North J.P., Harirchian P., Hailer A.A., Zhao Z., Ghadially R., et al. Classification of human chronic inflammatory skin disease based on single-cell immune profiling. Sci. Immunol. 2022;7:eabl9165. doi: 10.1126/sciimmunol.abl9165. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

