Exploring the Antiangiogenic and Anti-Inflammatory Potential of Homoisoflavonoids: Target Identification Using Biotin Probes
- PMID: 39062499
- PMCID: PMC11274659
- DOI: 10.3390/biom14070785
Exploring the Antiangiogenic and Anti-Inflammatory Potential of Homoisoflavonoids: Target Identification Using Biotin Probes
Abstract
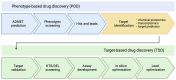
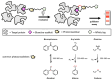
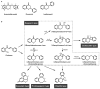
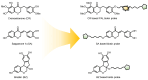
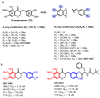
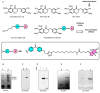
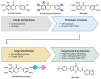
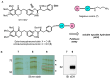
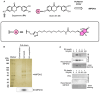
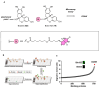
Chemical proteomics using biotin probes of natural products have significantly advanced our understanding of molecular targets and therapeutic potential. This review highlights recent progress in the application of biotin probes of homoisoflavonoids for identifying binding proteins and elucidating mechanisms of action. Notably, homoisoflavonoids exhibit antiangiogenic, anti-inflammatory, and antidiabetic effects. A combination of biotin probes, pull-down assays, mass spectrometry, and molecular modeling has revealed how natural products and their derivatives interact with several proteins such as ferrochelatase (FECH), soluble epoxide hydrolase (sEH), inosine monophosphate dehydrogenase 2 (IMPDH2), phosphodiesterase 4 (PDE4), and deoxyhypusine hydroxylase (DOHH). These target identification approaches pave the way for new therapeutic avenues, especially in the fields of oncology and ophthalmology. Future research aimed at expanding the repertoire of target identification using biotin probes of homoisoflavonoids promises to further elucidate the complex mechanisms and develop new drug candidates.
Keywords: homoisoflavonoid; natural products; photoaffinity labeling; target identification.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

