Enzymatic Transglycosylation Features in Synthesis of 8-Aza-7-Deazapurine Fleximer Nucleosides by Recombinant E. coli PNP: Synthesis and Structure Determination of Minor Products
- PMID: 39062512
- PMCID: PMC11275124
- DOI: 10.3390/biom14070798
Enzymatic Transglycosylation Features in Synthesis of 8-Aza-7-Deazapurine Fleximer Nucleosides by Recombinant E. coli PNP: Synthesis and Structure Determination of Minor Products
Abstract
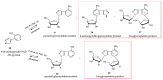
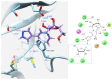
Enzymatic transglycosylation of the fleximer base 4-(4-aminopyridine-3-yl)-1H-pyrazole using recombinant E. coli purine nucleoside phosphorylase (PNP) resulted in the formation of "non-typical" minor products of the reaction. In addition to "typical" N1-pyrazole nucleosides, a 4-imino-pyridinium riboside and a N1-pyridinium-N1-pyrazole bis-ribose derivative were formed. N1-Pyrazole 2'-deoxyribonucleosides and a N1-pyridinium-N1-pyrazole bis-2'-deoxyriboside were formed. But 4-imino-pyridinium deoxyriboside was not formed in the reaction mixture. The role of thermodynamic parameters of key intermediates in the formation of reaction products was elucidated. To determine the mechanism of binding and activation of heterocyclic substrates in the E. coli PNP active site, molecular modeling of the fleximer base and reaction products in the enzyme active site was carried out. As for N1-pyridinium riboside, there are two possible locations for it in the PNP active site. The presence of a relatively large space in the area of amino acid residues Phe159, Val178, and Asp204 allows the ribose residue to fit into that space, and the heterocyclic base can occupy a position that is suitable for subsequent glycosylation. Perhaps it is this "upside down" arrangement that promotes secondary glycosylation and the formation of minor bis-riboside products.
Keywords: catalytic site; fleximer base; molecular modeling; recombinant E. coli PNP.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


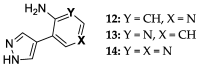








References
-
- Rinaldi F., Fernández-Lucas J., de La Fuente D., Zheng C., Bavaro T., Peters B., Massolini G., Annunziata F., Conti P., de la Mata I., et al. Immobilized enzyme reactors based on nucleoside phosphorylases and 2′-deoxyribosyltransferase for the in-flow synthesis of pharmaceutically relevant nucleoside analogues. Bioresour. Technol. 2020;307:123258. doi: 10.1016/j.biortech.2020.123258. - DOI - PubMed
-
- Calleri E., Cattaneo G., Rabuffetti M., Serra I., Bavaro T., Massolini G., Speranza G., Ubiali D. Flow-synthesis of nucleosides catalyzed by an immobilized purine nucleoside phosphorylase from Aeromonas hydrophila: Integrated systems of reaction control and product purification. Adv. Synth. Catal. 2015;357:2520–2528. doi: 10.1002/adsc.201500133. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

