Calreticulin-Enigmatic Discovery
- PMID: 39062580
- PMCID: PMC11275038
- DOI: 10.3390/biom14070866
Calreticulin-Enigmatic Discovery
Abstract
Calreticulin (CRT) is an intrinsically disordered multifunctional protein that plays essential roles intra-and extra-cellularly. The Michalak laboratory has proposed that CRT was initially identified in 1974 by the MacLennan laboratory as the high-affinity Ca2+-binding protein (HACBP) of the sarcoplasmic reticulin (SR). This widely accepted belief has been ingrained in the scientific literature but has never been rigorously tested. In our report, we have undertaken a comprehensive reexamination of this assumption by meticulously examining the majority of published studies that present a proteomic analysis of the SR. These analyses have utilized proteomic analysis of purified SR preparations or purified components of the SR, namely the longitudinal tubules and junctional terminal cisternae. These studies have consistently failed to detect the HACBP or CRT in skeletal muscle SR. We propose that the existence of the HACBP has failed the test of reproducibility and should be retired to the annals of antiquity. Therefore, the scientific dogma that the HACBP and CRT are identical proteins is a non sequitur.
Keywords: Michalak; calregulin; calreticulin; calsequestrin; endoplasmic reticulum; essential thrombocythemia; high-affinity calcium-binding protein (HACBP); sarcoplasmic reticulum.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




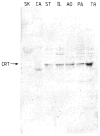






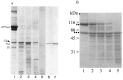
References
-
- MacLennan D.H., Yip C.C., Iles G.H., Seeman P. Cold Spring Harbor Symposia on Quantitative Biology. Volume 37. Cold Spring Harbor Laboratory Press; Long Island, NY, USA: 1973. Isolation of Sarcoplasmic Reticulum Proteins; pp. 469–477. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

