Identification and Comprehensive Analysis of OFP Genes for Fruit Shape Influence in Mango
- PMID: 39062602
- PMCID: PMC11275924
- DOI: 10.3390/genes15070823
Identification and Comprehensive Analysis of OFP Genes for Fruit Shape Influence in Mango
Abstract
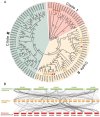
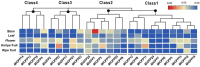
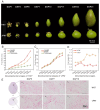
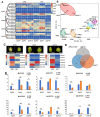
OVATE family proteins (OFPs) are a class of plant-specific proteins with a conserved OVATE domain that play fundamental roles in fruit development and plant growth. Mango (Mangifera indica L.) is an economically important subtropical fruit tree characterized by a diverse array of fruit shapes and sizes. Despite extensive research on OFPs across various species, there remains a scarcity of information regarding OFPs in mango. Here, we have successfully identified 25 OFP genes (MiOFPs) in mango, each of which exhibits the conserved OVATE domains. The MiOFP gene exhibit a range of 2-6 motifs, with all genes containing both motif 1 and motif 2. Phylogenetic analysis on 97 OFPs (including 18 AtOFPs, 24 SlOFPs, 25 MiOFPs, and 30 OsOFPs) indicated that MiOFPs could be divided into three main clades: clade I, II, and III. Comparative morphological analysis identified significant variations in fruit longitudinal diameter, fruit transverse diameter, and fruit shape index between two distinct shaped mango cultivars ('Hongxiangya' and 'Jingpingmang') at DAP5, DAP7, and DAP10 stages. The subsequent examination of paraffin sections revealed distinct patterns of cell elongation. The majority of MiOFP genes exhibited predominantly expressed in developing organs, specifically flowers and immature fruits, while displaying distinct expression patterns. RNA-Seq analysis revealed significant disparities in the expression levels of several OFP genes, including MiOFP5, MiOFP11, MiOFP21, MiOFP22, MiOFP23, and MiOFP25, between the two mango cultivars. These findings suggest that these six genes may play a crucial role for fruit shape in mango, especially the MiOFP22. The findings of this study have established a basis for future investigations into MiOFPs in mango, offering a solid foundation for further research in this field.
Keywords: OVATE family proteins; expression pattern; fruit development; mango (Mangifera indica L.).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Zhang C., Xie D., Bai T., Luo X., Zhang F., Ni Z., Chen Y. Diversity of a Large Collection of Natural Populations of Mango (Mangifera indica Linn.) Revealed by Agro-Morphological and Quality Traits. Diversity. 2020;12:27. doi: 10.3390/d12010027. - DOI
-
- Kulkarni M.M., Burondkar M.M., Dalvi N.V., Salvi B.R., Haldankar P.M., Bhattacharyya T. Mango Fruit Size Diversity found in Konkan. Adv. Agric. Res. Technol. J. 2019;3:43–46.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

