Genome-Wide Identification of the Whirly Gene Family and Its Potential Function in Low Phosphate Stress in Soybean (Glycine max)
- PMID: 39062612
- PMCID: PMC11275625
- DOI: 10.3390/genes15070833
Genome-Wide Identification of the Whirly Gene Family and Its Potential Function in Low Phosphate Stress in Soybean (Glycine max)
Abstract
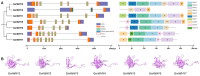
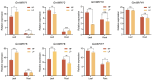
The Whirly (WHY) gene family, functioning as transcription factors, plays an essential role in the regulation of plant metabolic responses, which has been demonstrated across multiple species. However, the WHY gene family and its functions in soybean remains unclear. In this paper, we conducted genome-wide screening and identification to characterize the WHY gene family. Seven WHY members were identified and randomly distributed across six chromosomes. The phylogenetic evolutionary tree of WHY genes in soybean and other species was divided into five clades. An in-depth analysis revealed that segmental duplications significantly contributed to the expansion of GmWHYs, and the GmWHY gene members may have experienced evolutionary pressure for purifying selection in soybeans. The analysis of promoter Cis-elements in GmWHYs suggested their potential significance in addressing diverse stress conditions. The expression patterns of GmWHYs exhibited tissue-specific variations throughout the different stages of soybean development. Additionally, six GmWHY genes exhibited different responses to low phosphate stress. These findings will provide a theoretical basis and valuable reference for the future exploration of WHY gene function.
Keywords: expansion; expression patterns; haplotype; phosphate deficiency; whirly.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Genome-wide identification of MGT gene family in soybean (Glycine max) and their expression analyses under magnesium stress conditions.BMC Plant Biol. 2025 Jan 22;25(1):83. doi: 10.1186/s12870-024-05985-7. BMC Plant Biol. 2025. PMID: 39838318 Free PMC article.
-
Genome-Wide Analysis of AGC Genes Related to Salt Stress in Soybeans (Glycine max).Int J Mol Sci. 2025 Mar 13;26(6):2588. doi: 10.3390/ijms26062588. Int J Mol Sci. 2025. PMID: 40141228 Free PMC article.
-
Genome-wide analysis of filamentous temperature-sensitive H protease (ftsH) gene family in soybean.BMC Genomics. 2024 May 27;25(1):524. doi: 10.1186/s12864-024-10389-w. BMC Genomics. 2024. PMID: 38802777 Free PMC article.
-
Genome-wide identification, evolution, and expression analysis of carbonic anhydrases genes in soybean (Glycine max).Funct Integr Genomics. 2023 Jan 14;23(1):37. doi: 10.1007/s10142-023-00966-9. Funct Integr Genomics. 2023. PMID: 36639600 Review.
-
Genome-wide identification and expression analysis of the Dof gene family reveals their involvement in hormone response and abiotic stresses in sunflower (Helianthus annuus L.).Gene. 2024 Jun 5;910:148336. doi: 10.1016/j.gene.2024.148336. Epub 2024 Mar 4. Gene. 2024. PMID: 38447680 Review.
Cited by
-
Genome-Wide Identification and Comprehensive Analysis of the PPO Gene Family in Glycine max and Glycine soja.Genes (Basel). 2024 Dec 26;16(1):17. doi: 10.3390/genes16010017. Genes (Basel). 2024. PMID: 39858564 Free PMC article.
References
-
- Golin S., Negroni Y.L., Bennewitz B., Klosgen R.B., Mulisch M., La Rocca N., Cantele F., Vigani G., Lo Schiavo F., Krupinska K., et al. WHIRLY2 plays a key role in mitochondria morphology, dynamics, and functionality in Arabidopsis thaliana. Plant Direct. 2020;4:e00229. doi: 10.1002/pld3.229. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

