Comparative Analysis and Phylogeny of the Complete Chloroplast Genomes of Nine Cynanchum (Apocynaceae) Species
- PMID: 39062662
- PMCID: PMC11275380
- DOI: 10.3390/genes15070884
Comparative Analysis and Phylogeny of the Complete Chloroplast Genomes of Nine Cynanchum (Apocynaceae) Species
Abstract
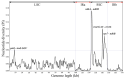
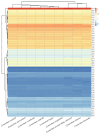
Cynanchum belongs to the Apocynaceae family and is a morphologically diverse genus that includes around 200 shrub or perennial herb species. Despite the utilization of CPGs, few molecular phylogenetic studies have endeavored to elucidate infrafamilial relationships within Cynanchum through extensive taxon sampling. In this research, we constructed a phylogeny and estimated divergence time based on the chloroplast genomes (CPGs) of nine Cynanchum species. We sequenced and annotated nine chloroplast (CP) genomes in this study. The comparative analysis of these genomes from these Cynanchum species revealed a typical quadripartite structure, with a total sequence length ranging from 158,283 to 161,241 base pairs (bp). The CP genome (CPG) was highly conserved and moderately differentiated. Through annotation, we identified a total of 129-132 genes. Analysis of the boundaries of inverted repeat (IR) regions showed consistent positioning: the rps19 gene was located in the IRb region, varying from 46 to 50 bp. IRb/SSC junctions were located between the trnN and ndhF genes. We did not detect major expansions or contractions in the IR region or rearrangements or insertions in the CPGs of the nine Cynanchum species. The results of SSR analysis revealed a variation in the number of SSRs, ranging from 112 to 150. In five types of SSRs, the largest number was mononucleotide repeats, and the smallest number was hexanucleotide repeats. The number of long repeats in the cp genomes of nine Cynanchum species was from 35 to 80. In nine species of Cynanchum, the GC3s values ranged from 26.80% to 27.00%, indicating a strong bias towards A/U-ending codons. Comparative analyses revealed four hotspot regions in the CPG, ndhA-ndhH, trnI-GAU-rrn16, psbI-trnS-GCU, and rps7-ndhB, which could potentially serve as molecular markers. In addition, phylogenetic tree construction based on the CPG indicated that the nine Cynanchum species formed a monophyletic group. Molecular dating suggested that Cynanchum diverged from its sister genus approximately 18.87 million years ago (Mya) and species diversification within the Cynanchum species primarily occurred during the recent Miocene epoch. The divergence time estimation presented in this study will facilitate future research on Cynanchum, aid in species differentiation, and facilitate diverse investigations into this economically and ecologically important genus.
Keywords: Cynanchum; chloroplast genomes; comparative analysis; phylogenomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Li P.T., Michael G.G., Stevens W.D. Asclepiadaceae. In: Wu Z., Raven P., editors. Flora of China. Volume 16. Science Press; Beijing, China: 1995. pp. 311–313.
-
- Endress M.E., Liede-Schumann S., Meve U. An updated classification for Apocynaceae. Phytotaxa. 2014;159:115. doi: 10.11646/phytotaxa.159.3.2. - DOI
-
- Yang J., Qing Z., Wang D., Yang H. Distribution of medicinal plant resources of Cynanchum in Guizhou. Guizhou Sci. 2018;36:50–52.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

