New Virus Variant Detection Based on the Optimal Natural Metric
- PMID: 39062670
- PMCID: PMC11275827
- DOI: 10.3390/genes15070891
New Virus Variant Detection Based on the Optimal Natural Metric
Abstract
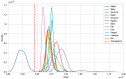
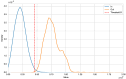
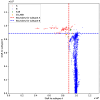
The highly variable SARS-CoV-2 virus responsible for the COVID-19 pandemic frequently undergoes mutations, leading to the emergence of new variants that present novel threats to public health. The determination of these variants often relies on manual definition based on local sequence characteristics, resulting in delays in their detection relative to their actual emergence. In this study, we propose an algorithm for the automatic identification of novel variants. By leveraging the optimal natural metric for viruses based on an alignment-free perspective to measure distances between sequences, we devise a hypothesis testing framework to determine whether a given viral sequence belongs to a novel variant. Our method demonstrates high accuracy, achieving nearly 100% precision in identifying new variants of SARS-CoV-2 and HIV-1 as well as in detecting novel genera in Orthocoronavirinae. This approach holds promise for timely surveillance and management of emerging viral threats in the field of public health.
Keywords: SARS-CoV-2; natural vectors; new virus detection; optimal metric.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study, in the collection, analysis, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures
References
-
- Uddin M., Mustafa F., Rizvi T.A., Loney T., Al Suwaidi H., Al-Marzouqi A.H.H., Kamal Eldin A., Alsabeeha N., Adrian T.E., Stefanini C., et al. SARS-CoV-2/COVID-19: Viral Genomics, Epidemiology, Vaccines, and Therapeutic Interventions. Viruses. 2020;12:526. doi: 10.3390/v12050526. - DOI - PMC - PubMed
-
- Vulturar D.-M., Moacă L.-Ș., Neag M.A., Mitre A.-O., Alexescu T.-G., Gherman D., Făgărășan I., Chețan I.M., Gherman C.D., Melinte O.-E., et al. Delta Variant in the COVID-19 Pandemic: A Comparative Study on Clinical Outcomes Based on Vaccination Status. J. Pers. Med. 2024;14:358. doi: 10.3390/jpm14040358. - DOI - PMC - PubMed
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous