Sulfotransferase 4A1 Coding Sequence and Protein Structure Are Highly Conserved in Vertebrates
- PMID: 39062693
- PMCID: PMC11275347
- DOI: 10.3390/genes15070914
Sulfotransferase 4A1 Coding Sequence and Protein Structure Are Highly Conserved in Vertebrates
Abstract
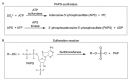
Cytosolic sulfotransferases (SULTs) are Phase 2 drug-metabolizing enzymes that catalyze the conjugation of sulfonate to endogenous and xenobiotic compounds, increasing their hydrophilicity and excretion from cells. To date, 13 human SULTs have been identified and classified into five families. SULT4A1 mRNA encodes two variants: (1) the wild type, encoding a 284 amino acid, ~33 kDa protein, and (2) an alternative spliced variant resulting from a 126 bp insert between exon 6 and 7, which introduces a premature stop codon that enhances nonsense-mediated decay. SULT4A1 is classified as an SULT based on sequence and structural similarities, including PAPS-domains, active-site His, and the dimerization domain; however, the catalytic pocket lid 'Loop 3' size is not conserved. SULT4A1 is uniquely expressed in the brain and localized in the cytosol and mitochondria. SULT4A1 is highly conserved, with rare intronic polymorphisms that have no outward manifestations. However, the SULT4A1 haplotype is correlated with Phelan-McDermid syndrome and schizophrenia. SULT4A1 knockdown revealed potential SULT4A1 functions in photoreceptor signaling and knockout mice display hampered neuronal development and behavior. Mouse and yeast models revealed that SULT4A1 protects the mitochondria from endogenously and exogenously induced oxidative stress and stimulates cell division, promoting dendritic spines' formation and synaptic transmission. To date, no physiological enzymatic activity has been associated with SULT4A1.
Keywords: gene structure; polymorphism; protein structure; tissue/cell distribution.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






Similar articles
-
Sulfotransferase 4A1 activity facilitates sulfate-dependent cellular protection to oxidative stress.Sci Rep. 2022 Jan 31;12(1):1625. doi: 10.1038/s41598-022-05582-4. Sci Rep. 2022. PMID: 35102205 Free PMC article.
-
SULT4A1 Modulates Synaptic Development and Function by Promoting the Formation of PSD-95/NMDAR Complex.J Neurosci. 2020 Sep 9;40(37):7013-7026. doi: 10.1523/JNEUROSCI.2194-19.2020. Epub 2020 Aug 12. J Neurosci. 2020. PMID: 32801157 Free PMC article.
-
Generation and Characterization of SULT4A1 Mutant Mouse Models.Drug Metab Dispos. 2018 Jan;46(1):41-45. doi: 10.1124/dmd.117.077560. Epub 2017 Nov 6. Drug Metab Dispos. 2018. PMID: 29109113 Free PMC article.
-
Sulfotransferase 4A1.Int J Biochem Cell Biol. 2008;40(12):2686-91. doi: 10.1016/j.biocel.2007.11.010. Epub 2007 Dec 3. Int J Biochem Cell Biol. 2008. PMID: 18248844 Review.
-
Structural plasticity in the human cytosolic sulfotransferase dimer and its role in substrate selectivity and catalysis.Drug Metab Pharmacokinet. 2015 Feb;30(1):3-20. doi: 10.1016/j.dmpk.2014.10.004. Epub 2014 Nov 5. Drug Metab Pharmacokinet. 2015. PMID: 25760527 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

