Exploring Microorganisms Associated to Acute Febrile Illness and Severe Neurological Disorders of Unknown Origin: A Nanopore Metagenomics Approach
- PMID: 39062701
- PMCID: PMC11276239
- DOI: 10.3390/genes15070922
Exploring Microorganisms Associated to Acute Febrile Illness and Severe Neurological Disorders of Unknown Origin: A Nanopore Metagenomics Approach
Abstract
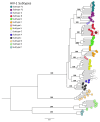
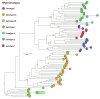
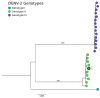
Acute febrile illness (AFI) and severe neurological disorders (SNDs) often present diagnostic challenges due to their potential origins from a wide range of infectious agents. Nanopore metagenomics is emerging as a powerful tool for identifying the microorganisms potentially responsible for these undiagnosed clinical cases. In this study, we aim to shed light on the etiological agents underlying AFI and SND cases that conventional diagnostic methods have not been able to fully elucidate. Our approach involved analyzing samples from fourteen hospitalized patients using a comprehensive nanopore metagenomic approach. This process included RNA extraction and enrichment using the SMART-9N protocol, followed by nanopore sequencing. Subsequent steps involved quality control, host DNA/cDNA removal, de novo genome assembly, and taxonomic classification. Our findings in AFI cases revealed a spectrum of disease-associated microbes, including Escherichia coli, Streptococcus sp., Human Immunodeficiency Virus 1 (Subtype B), and Human Pegivirus. Similarly, SND cases revealed the presence of pathogens such as Escherichia coli, Clostridium sp., and Dengue virus type 2 (Genotype-II lineage). This study employed a metagenomic analysis method, demonstrating its efficiency and adaptability in pathogen identification. Our investigation successfully identified pathogens likely associated with AFI and SNDs, underscoring the feasibility of retrieving near-complete genomes from RNA viruses. These findings offer promising prospects for advancing our understanding and control of infectious diseases, by facilitating detailed genomic analysis which is critical for developing targeted interventions and therapeutic strategies.
Keywords: acute febrile illness; metagenomics; nanopore sequencing; severe neurological disorders.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Bressan C.D.S., Teixeira M.L.B., Gouvêa M.I.F.D.S., de Pina-Costa A., Santos H.F.P., Calvet G.A., Lupi O., Siqueira A.M., Valls-de-Souza R., Valim C., et al. Challenges of acute febrile illness diagnosis in a national infectious diseases center in Rio de Janeiro: 16-year experience of syndromic surveillance. PLoS Negl. Trop. Dis. 2023;17:e0011232. doi: 10.1371/journal.pntd.0011232. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

