Reprogramming of Glutamine Amino Acid Transporters Expression and Prognostic Significance in Hepatocellular Carcinoma
- PMID: 39062801
- PMCID: PMC11277143
- DOI: 10.3390/ijms25147558
Reprogramming of Glutamine Amino Acid Transporters Expression and Prognostic Significance in Hepatocellular Carcinoma
Abstract
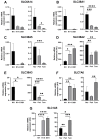
Hepatocellular carcinoma (HCC) is the most prevalent primary liver malignancy and is a major cause of cancer-related mortality in the world. This study aimed to characterize glutamine amino acid transporter expression profiles in HCC compared to those of normal liver cells. In vitro and in vivo models of HCC were studied using qPCR, whereas the prognostic significance of glutamine transporter expression levels within patient tumors was analyzed through RNAseq. Solute carrier (SLC) 1A5 and SLC38A2 were targeted through siRNA or gamma-p-nitroanilide (GPNA). HCC cells depended on exogenous glutamine for optimal survival and growth. Murine HCC cells showed superior glutamine uptake rate than normal hepatocytes (p < 0.0001). HCC manifested a global reprogramming of glutamine transporters compared to normal liver: SLC38A3 levels decreased, whereas SLC38A1, SLC7A6, and SLC1A5 levels increased. Also, decreased SLC6A14 and SLC38A3 levels or increased SLC38A1, SLC7A6, and SLC1A5 levels predicted worse survival outcomes (all p < 0.05). Knockdown of SLC1A5 and/or SLC38A2 expression in human Huh7 and Hep3B HCC cells, as well as GPNA-mediated inhibition, significantly decreased the uptake of glutamine; combined SLC1A5 and SLC38A2 targeting had the most considerable impact (all p < 0.05). This study revealed glutamine transporter reprogramming as a novel hallmark of HCC and that such expression profiles are clinically significant.
Keywords: glutamine; hepatocellular carcinoma; liver; metabolic reprogramming; transporters.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures





Similar articles
-
LncRNA SLC1A5-AS/MZF1/ASCT2 Axis Contributes to Malignant Progression of Hepatocellular Carcinoma.Discov Med. 2023 Dec;35(179):995-1014. doi: 10.24976/Discov.Med.202335179.96. Discov Med. 2023. PMID: 38058065
-
Elevated SLC1A5 associated with poor prognosis and therapeutic resistance to transarterial chemoembolization in hepatocellular carcinoma.J Transl Med. 2024 Jun 6;22(1):543. doi: 10.1186/s12967-024-05298-1. J Transl Med. 2024. PMID: 38844930 Free PMC article.
-
The tRNA Gm18 methyltransferase TARBP1 promotes hepatocellular carcinoma progression via metabolic reprogramming of glutamine.Cell Death Differ. 2024 Sep;31(9):1219-1234. doi: 10.1038/s41418-024-01323-4. Epub 2024 Jun 12. Cell Death Differ. 2024. PMID: 38867004 Free PMC article.
-
The transport of glutamine into mammalian cells.Front Biosci. 2007 Jan 1;12:874-82. doi: 10.2741/2109. Front Biosci. 2007. PMID: 17127344 Review.
-
Pathophysiological role of ion channels and transporters in hepatocellular carcinoma.Cancer Gene Ther. 2024 Nov;31(11):1611-1618. doi: 10.1038/s41417-024-00782-8. Epub 2024 Jul 24. Cancer Gene Ther. 2024. PMID: 39048663 Free PMC article. Review.
Cited by
-
Targeting glutamine metabolism as a potential target for cancer treatment.J Exp Clin Cancer Res. 2025 Jul 1;44(1):180. doi: 10.1186/s13046-025-03430-7. J Exp Clin Cancer Res. 2025. PMID: 40598593 Free PMC article. Review.
-
Analysis of novel therapeutic targets and construction of a prognostic model for hepatocellular carcinoma.PeerJ. 2025 Aug 22;13:e19899. doi: 10.7717/peerj.19899. eCollection 2025. PeerJ. 2025. PMID: 40860678 Free PMC article.
References
-
- Global Burden of Disease Cancer Collaboration. Fitzmaurice C., Allen C., Barber R.M., Barregard L., Bhutta Z.A., Brenner H., Dicker D.J., Chimed-Orchir O., Dandona R., et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

