Mechanism of Histone Arginine Methylation Dynamic Change in Cellular Stress
- PMID: 39062806
- PMCID: PMC11277302
- DOI: 10.3390/ijms25147562
Mechanism of Histone Arginine Methylation Dynamic Change in Cellular Stress
Abstract
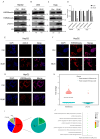
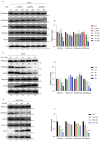
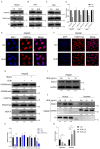
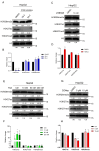
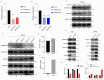
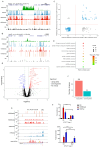
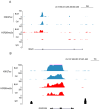
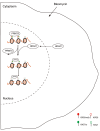
Histone arginine residue methylation is crucial for individual development and gene regulation. However, the dynamics of histone arginine methylation in response to cellular stress remains largely unexplored. In addition, the interplay and regulatory mechanisms between this and other histone modifications are important scientific questions that require further investigation. This study aimed to investigate the changes in histone arginine methylation in response to DNA damage. We report a global decrease in histone H3R26 symmetric dimethylation (H3R26me2s) and hypoacetylation at the H3K27 site in response to DNA damage. Notably, H3R26me2s exhibits a distribution pattern similar to that of H3K27ac across the genome, both of which are antagonistic to H3K27me3. Additionally, histone deacetylase 1 (HDAC1) may be recruited to the H3R26me2s demethylation region to mediate H3K27 deacetylation. These findings suggest crosstalk between H3R26me2s and H3K27ac in regulating gene expression.
Keywords: H3K27ac; H3R26me2s; crosstalk; stress.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures

References
-
- Cheng Y., Wang X., Huang S., Zhang L., Lan B., Li X., Chen H., Liu Z., Su Y., Xi L., et al. A CRISPR-Cas9 library screening identifies CARM1 as a critical inhibitor of ferroptosis in hepatocellular carcinoma cells. Mol. Ther. Nucleic Acids. 2023;34:102063. doi: 10.1016/j.omtn.2023.102063. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

