CryoEM Workflow Acceleration with Feret Signatures
- PMID: 39062836
- PMCID: PMC11277255
- DOI: 10.3390/ijms25147593
CryoEM Workflow Acceleration with Feret Signatures
Abstract
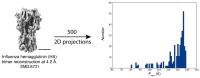
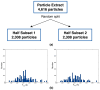
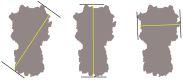
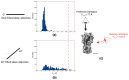
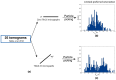
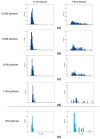
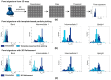
Common challenges in cryogenic electron microscopy, such as orientation bias, conformational diversity, and 3D misclassification, complicate single particle analysis and lead to significant resource expenditure. We previously introduced an in silico method using the maximum Feret diameter distribution, the Feret signature, to characterize sample heterogeneity of disc-shaped samples. Here, we expanded the Feret signature methodology to identify preferred orientations of samples containing arbitrary shapes with only about 1000 particles required. This method enables real-time adjustments of data acquisition parameters for optimizing data collection strategies or aiding in decisions to discontinue ineffective imaging sessions. Beyond detecting preferred orientations, the Feret signature approach can serve as an early-warning system for inconsistencies in classification during initial image processing steps, a capability that allows for strategic adjustments in data processing. These features establish the Feret signature as a valuable auxiliary tool in the context of single particle analysis, significantly accelerating the structure determination process.
Keywords: Feret signature; analytical tool; cryogenic electron microscopy; data processing; single particle analysis; structural biology; structure determination; workflow optimization.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Multi-species cryoEM calibration and workflow verification standard.Acta Crystallogr F Struct Biol Commun. 2024 Nov 1;80(Pt 11):320-327. doi: 10.1107/S2053230X24010318. Epub 2024 Oct 31. Acta Crystallogr F Struct Biol Commun. 2024. PMID: 39494502 Free PMC article.
-
Algorithmic robustness to preferred orientations in single particle analysis by CryoEM.J Struct Biol. 2021 Mar;213(1):107695. doi: 10.1016/j.jsb.2020.107695. Epub 2021 Jan 7. J Struct Biol. 2021. PMID: 33421545
-
Characterization of heterogeneity in nanodisc samples using Feret signatures.J Struct Biol. 2022 Dec;214(4):107916. doi: 10.1016/j.jsb.2022.107916. Epub 2022 Nov 1. J Struct Biol. 2022. PMID: 36332745
-
Emerging Themes in CryoEM─Single Particle Analysis Image Processing.Chem Rev. 2022 Sep 14;122(17):13915-13951. doi: 10.1021/acs.chemrev.1c00850. Epub 2022 Jul 4. Chem Rev. 2022. PMID: 35785962 Free PMC article. Review.
-
Tools for Model Building and Optimization into Near-Atomic Resolution Electron Cryo-Microscopy Density Maps.Methods Enzymol. 2016;579:255-76. doi: 10.1016/bs.mie.2016.06.003. Epub 2016 Aug 12. Methods Enzymol. 2016. PMID: 27572730 Free PMC article. Review.
Cited by
-
Optimizing Transmembrane Protein Assemblies in Nanodiscs for Structural Studies: A Comprehensive Manual.Bio Protoc. 2024 Nov 5;14(21):e5099. doi: 10.21769/BioProtoc.5099. eCollection 2024 Nov 5. Bio Protoc. 2024. PMID: 39525973 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

