Conserved Signaling Pathways in the Ciona robusta Gut
- PMID: 39063090
- PMCID: PMC11277035
- DOI: 10.3390/ijms25147846
Conserved Signaling Pathways in the Ciona robusta Gut
Abstract
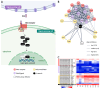
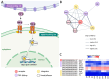
The urochordate Ciona robusta exhibits numerous functional and morphogenetic traits that are shared with vertebrate models. While prior investigations have identified several analogies between the gastrointestinal tract (i.e., gut) of Ciona and mice, the molecular mechanisms responsible for these similarities remain poorly understood. This study seeks to address this knowledge gap by investigating the transcriptional landscape of the adult stage gut. Through comparative genomics analyses, we identified several evolutionarily conserved components of signaling pathways of pivotal importance for gut development (such as WNT, Notch, and TGFβ-BMP) and further evaluated their expression in three distinct sections of the gastrointestinal tract by RNA-seq. Despite the presence of lineage-specific gene gains, losses, and often unclear orthology relationships, the investigated pathways were characterized by well-conserved molecular machinery, with most components being expressed at significant levels throughout the entire intestinal tract of C. robusta. We also showed significant differences in the transcriptional landscape of the stomach and intestinal tract, which were much less pronounced between the proximal and distal portions of the intestine. This study confirms that C. robusta is a reliable model system for comparative studies, supporting the use of ascidians as a model to study gut physiology.
Keywords: Ciona robusta; RNAseq; digestive tract; gut; signaling pathway; urochordate.
Conflict of interest statement
The authors declare they have no competing financial or non-financial interests.
Figures



References
-
- Wanninger A. Evolutionary Developmental Biology of Invertebrates 6: Deuterostomia. Springer; Berlin, Germany: 2015.
-
- Burighel P., Cloney R.A. Microscopic Anatomy of Invertebrates, Volume 15: Hemichordata, Chaetognata, and Invertebrate Chordates. Wiley-Liss; New York, NY, USA: 1997. Urochordata, Ascidiacea; pp. 221–347.
-
- Liberti A., Bertocci I., Pollet A., Musco L., Locascio A., Ristoratore F., Spagnuolo A., Sordino P. An Indoor Study of the Combined Effect of Industrial Pollution and Turbulence Events on the Gut Environment in a Marine Invertebrate: The Response of the Gut Environment to Polluted Sediment Reworking. Mar. Environ. Res. 2020;158:104950. doi: 10.1016/j.marenvres.2020.104950. - DOI - PubMed
-
- Caputi L., Toscano F., Arienzo M., Ferrara L., Procaccini G., Sordino P. Temporal Correlation of Population Composition and Environmental Variables in the Marine Invader Ciona Robusta. Mar. Ecol. 2019;40:e12543. doi: 10.1111/maec.12543. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

