Comparative Analysis of Plastome Sequences of Seven Tulipa L. (Liliaceae Juss.) Species from Section Kolpakowskianae Raamsd. Ex Zonn and Veldk
- PMID: 39063115
- PMCID: PMC11277319
- DOI: 10.3390/ijms25147874
Comparative Analysis of Plastome Sequences of Seven Tulipa L. (Liliaceae Juss.) Species from Section Kolpakowskianae Raamsd. Ex Zonn and Veldk
Abstract
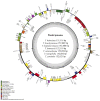
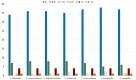
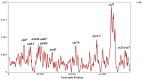
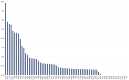
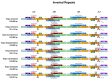
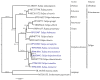
Tulipa L. is a genus of significant economic, environmental, and cultural importance in several parts of the world. The exact number of species in the genus remains uncertain due to inherent taxonomic challenges. We utilized next-generation sequencing technology to sequence and assemble the plastid genomes of seven Tulipa species collected in Kazakhstan and conducted a comparative analysis. The total number of annotated genes was 136 in all seven studied Tulipa species, 114 of which were unique, including 80 protein-coding, 30 tRNA, and 4 rRNA genes. Nine regions (petD, ndhH, ycf2-ycf3, ndhA, rpl16, clpP, ndhD-ndhF, rpoC2, and ycf1) demonstrated significant nucleotide variability, suggesting their potential as molecular markers. A total of 1388 SSRs were identified in the seven Tulipa plastomes, with mononucleotide repeats being the most abundant (60.09%), followed by dinucleotide (34.44%), tetranucleotide (3.90%), trinucleotide (1.08%), pentanucleotide (0.22%), and hexanucleotide (0.29%). The Ka/Ks values of the protein-coding genes ranged from 0 to 3.9286, with the majority showing values <1. Phylogenetic analysis based on a complete plastid genome and protein-coding gene sequences divided the species into three major clades corresponding to their subgenera. The results obtained in this study may contribute to understanding the phylogenetic relationships and molecular taxonomy of Tulipa species.
Keywords: Liliaceae; Tulipa; next-generation sequencing; phylogenetic relationships; plastid genome; variable regions.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Veldkamp J.F., Zonneveld B.J.M. The infrageneric nomenclature of Tulipa (Liliaceae) Plant Syst. Evol. 2012;298:87–92. doi: 10.1007/s00606-011-0525-0. - DOI
-
- Ivaschenko A.A., Belyalov O.V. Kazakhstan is the Birthplace of Tulips. Atamura; Almaty, Kazakhstan: 2019.
-
- Botschantzeva Z.P. Tulips: Taxonomy, Morphology, Cytology, Phytogeography, and Physiology. CRC Press; Rotterdam, The Netherlands: 1962. (English Edition Translated by Varekamp, H.Q. 1982)
-
- Tojibaev K., Beshko N. Reassessment of diversity and analysis of distribution in Tulipa (Liliaceae) in Uzbekistan. Nord. J. Bot. 2015;33:324–334. doi: 10.1111/njb.00616. - DOI
-
- Beshko N.Y., Abduraimov O.S., Kodirov U.K., Madaminov F.M., Mahmudov A.V. The Current State of Cenopopulations of Some Endemic and Rare Species of the Genus Tulipa L. (Liliaceae) in the Tashkent Region (Uzbekistan) Arid Ecosyst. 2023;13:294–304. doi: 10.1134/S2079096123030034. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

