Genome-Wide Identification and Interaction Analysis of Turbot Heat Shock Protein 40 and 70 Families Suggest the Mechanism of Chaperone Proteins Involved in Immune Response after Bacterial Infection
- PMID: 39063205
- PMCID: PMC11277129
- DOI: 10.3390/ijms25147963
Genome-Wide Identification and Interaction Analysis of Turbot Heat Shock Protein 40 and 70 Families Suggest the Mechanism of Chaperone Proteins Involved in Immune Response after Bacterial Infection
Abstract
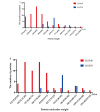
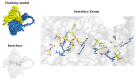
Hsp40-Hsp70 typically function in concert as molecular chaperones, and their roles in post-infection immune responses are increasingly recognized. However, in the economically important fish species Scophthalmus maximus (turbot), there is still a lack in the systematic identification, interaction models, and binding site analysis of these proteins. Herein, 62 Hsp40 genes and 16 Hsp70 genes were identified in the turbot at a genome-wide level and were unevenly distributed on 22 chromosomes through chromosomal distribution analysis. Phylogenetic and syntenic analysis provided strong evidence in supporting the orthologies and paralogies of these HSPs. Protein-protein interaction and expression analysis was conducted to predict the expression profile after challenging with Aeromonas salmonicida. dnajb1b and hspa1a were found to have a co-expression trend under infection stresses. Molecular docking was performed using Auto-Dock Tool and PyMOL for this pair of chaperone proteins. It was discovered that in addition to the interaction sites in the J domain, the carboxyl-terminal domain of Hsp40 also plays a crucial role in its interaction with Hsp70. This is important for the mechanistic understanding of the Hsp40-Hsp70 chaperone system, providing a theoretical basis for turbot disease resistance breeding, and effective value for the prevention of certain diseases in turbot.
Keywords: Aeromonas salmonicida; heat shock protein; molecular docking; turbot.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures







References
-
- Bjørndal T., Øiestad V. The Development of a New Farmed Species: Production Technology and Markets for Turbot. Institute for Research in Economics and Business Administration; Bergen, Norway: 2010. Working Paper.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

